













| General Information: |
|
| Name(s) found: |
DRP1B_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 610 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
GTP binding
[IEA][ISS]
GTPase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESLIALVNK IQRACTALGD HGEGSSLPTL WDSLPAIAVV GGQSSGKSSV LESVVGKDFL 60
61 PRGAGIVTRR PLVLQLHRID EGKEYAEFMH LPKKKFTDFA AVRQEISDET DRETGRSSKV 120
121 ISTVPIHLSI FSPNVVNLTL VDLPGLTKVA VDGQPESIVQ DIENMVRSFI EKPNCIILAI 180
181 SPANQDLATS DAIKISREVD PKGDRTFGVL TKIDLMDQGT NAVDILEGRG YKLRYPWVGV 240
241 VNRSQADINK SVDMIAARRR ERDYFQTSPE YRHLTERMGS EYLGKMLSKH LEVVIKSRIP 300
301 GLQSLITKTI SELETELSRL GKPVAADAGG KLYMIMEICR AFDQTFKEHL DGTRSGGEKI 360
361 NSVFDNQFPA AIKRLQFDKH LSMDNVRKLI TEADGYQPHL IAPEQGYRRL IESCLVSIRG 420
421 PAEAAVDAVH SILKDLIHKS MGETSELKQY PTLRVEVSGA AVDSLDRMRD ESRKATLLLV 480
481 DMESGYLTVE FFRKLPQDSE KGGNPTHSIF DRYNDAYLRR IGSNVLSYVN MVCAGLRNSI 540
541 PKSIVYCQVR EAKRSLLDIF FTELGQKEMS KLSKLLDEDP AVQQRRTSIA KRLELYRSAQ 600
601 TDIEAVAWSK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
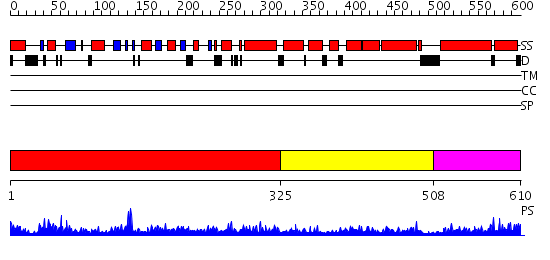
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..324] | 102.0 | Structure of the nucleotide-free myosin II motor domain from Dictyostelium discoideum fused to the GTPase domain of dynamin 1 from Rattus norvegicus | |
| 2 | View Details | [325..507] | 20.29243 | No description for PF01031.11 was found. No confident structure predictions are available. | |
| 3 | View Details | [508..610] | 32.79588 | No description for PF02212.9 was found. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)