













| General Information: |
|
| Name(s) found: |
gi|221234486
[NCBI NR]
gi|220963658 [NCBI NR] |
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Caulobacter crescentus NA1000 |
| Length: | 635 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAHQSALIAE DIEAYLHQHQ HKSLLRFITC GSVDDGKSTL IGRLLYDSKM IFEDQMAALE 60
61 VDSRKVGTQG GAIDFALLVD GLAAEREQGI TIDVAYRFFS TEKRKFIVAD TPGHEQYTRN 120
121 MVTGASTADA AVILIDARKG VLTQTRRHSY LVSLLGIRHV VLAVNKMDLV GWDKGVFDAI 180
181 VADYRAFAEQ IGLSVFTPIP ISGLGGDNMA TRSDATPWFE GPILMDWLEA VEVEDDLQAK 240
241 PFRMPVQWVN RPNLDFRGFS GLIASGSIKP GDRIRALPSG RESRVARIVT LPGDLDQAVA 300
301 GQSVTLTLED EIDISRGDVI AAADAPAPVA NQFEATLVWM DDEPLPPGRT YLLKLGARTV 360
361 GASITDIKHR VNVNTLEKTA AKRLELNEIG VCNLSLDQAI PFEAYAENRH MGGFILIDRL 420
421 SNRTVGAGLI NFALRRADNI HWQHTDVTKV SRAALKRQRG QVVWLTGLSG AGKSTIANLV 480
481 EKRLHALGRH TYLLDGDNVR HGLNKDLGFT EEDRVENIRR VAEVAKLMVD AGLIVLTAFI 540
541 SPFRAERQLA RDLLEPGEFI EVFVDTPLAV AEARDVKGLY KKARSGQLKN FTGVDSPYEA 600
601 PESPELRIDT TAIDPVEAAE RIVAWLEGQE IDYTI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
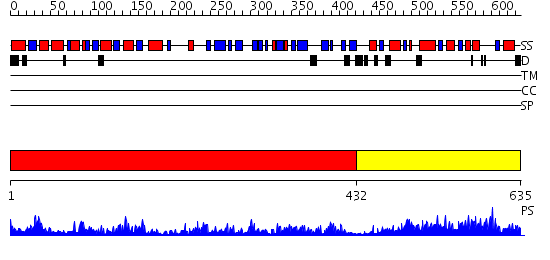
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..431] | 106.0 | Crystal Structure of a GTP-Regulated ATP Sulfurylase Heterodimer from Pseudomonas syringae | |
| 2 | View Details | [432..635] | 76.522879 | The crystal structure of human 3'-phosphoadenosine-5'-phosphosulfate synthetase 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)