













| General Information: |
|
| Name(s) found: |
gi|190407918
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 737 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALVKYSTVF FPLRSLRLFV SIKKAYYHSE PHSIDLFHDK DWIVKRPKFL NLPKNEHSKL 60
61 DIFQFNFNKS ESNNVYLQDS SFKDNLDKAM QFIYNDKLSS LDAKQVPIKN LAWLKLRDYI 120
121 YQQLKDPKLQ AKTYVPSVSE IIHPSSPGNL ISLLINCNKI SNLVWKSVLK YSLSNNITTL 180
181 DKFIHVLQQT FDHVYEQEIL PMMTNTDDTD GAHNVDITNP AEWFPEARKI RRHIIMHIGP 240
241 TNSGKTYRAL QKLKSVDRGY YAGPLRLLAR EVYDRFHAEK IRCNLLTGEE VIRDLDDRGN 300
301 SAGLTSGTVE MVPINQKFDV VVLDEIQMMS DGDRGWAWTN ALLGVVSKEV HLCGEKSVLP 360
361 LVKSIVKMTG DKLTINEYER LGKLSVEEKP IKDGIKGLRK GDCVVAFSKK KILDLKLKIE 420
421 KDTNLKVAVI YGSLPPETRV QQAALFNNGE YDIMVASDAI GMGLNLSIDR VVFTTNMKYN 480
481 GEELMEMTSS QIKQIGGRAG RFKSRSASGG VPQGFITSFE SKVLKSVRKA IEAPVEYLKT 540
541 AVTWPTDEIC AQLMTQFPPG TPTSVLLQTI SDELEKSSDN LFTLSDLKSK LKVIGLFEHM 600
601 EDIPFFDKLK LSNAPVKDMP MVTKAFTKFC ETIAKRHTRG LLSYRLPFNL LDYNCIPNES 660
661 YSLEVYESLY NIITLYFWLS NRYPNYFIDM ESAKDLKYFC EMIIFEKLDR LKKNPYAHKP 720
721 FGSTRGHLSS SRRRLRT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
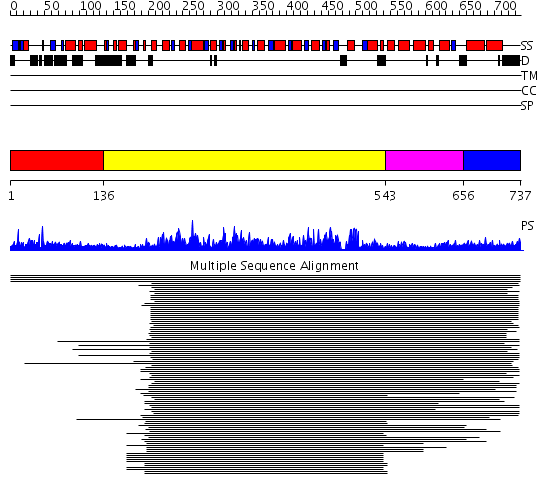
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..135] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [136..542] | 84.30103 | Initiation factor 4a | |
| 3 | View Details | [543..655] | 7.0 | Nucleotide excision repair enzyme UvrB | |
| 4 | View Details | [656..737] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)