













| General Information: |
|
| Name(s) found: |
CDPKW_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 538 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] plasma membrane [IDA] |
| Biological Process: |
abscisic acid mediated signaling pathway
[IMP]
response to salt stress [IEP] protein amino acid phosphorylation [ISS] |
| Molecular Function: |
protein binding
[IPI]
calmodulin-dependent protein kinase activity [ISS] kinase activity [ISS] calcium-dependent protein kinase C activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGNCCGTAGS LAQNDNKPKK GRKKQNPFSI DYGLHHGGGD GGGRPLKLIV LNDPTGREIE 60
61 SKYTLGRELG RGEFGVTYLC TDKETDDVFA CKSILKKKLR TAVDIEDVRR EVEIMRHMPE 120
121 HPNVVTLKET YEDEHAVHLV MELCEGGELF DRIVARGHYT ERAAAAVTKT IMEVVQVCHK 180
181 HGVMHRDLKP ENFLFGNKKE TAPLKAIDFG LSVFFKPGER FNEIVGSPYY MAPEVLKRNY 240
241 GPEVDIWSAG VILYILLCGV PPFWAETEQG VAQAIIRSVL DFRRDPWPKV SENAKDLIRK 300
301 MLDPDQKRRL TAQQVLDHPW LQNAKTAPNV SLGETVRARL KQFTVMNKLK KRALRVIAEH 360
361 LSDEEASGIR EGFQIMDTSQ RGKINIDELK IGLQKLGHAI PQDDLQILMD AGDIDRDGYL 420
421 DCDEFIAISV HLRKMGNDEH LKKAFAFFDQ NNNGYIEIEE LREALSDELG TSEEVVDAII 480
481 RDVDTDKDGR ISYEEFVTMM KTGTDWRKAS RQYSRERFNS ISLKLMQDAS LQVNGDTR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
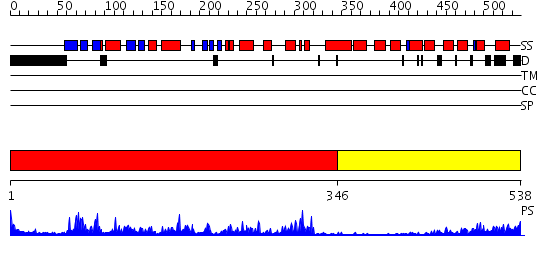
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..345] | 75.69897 | CALMODULIN-DEPENDENT PROTEIN KINASE FROM RAT | |
| 2 | View Details | [346..538] | 19.30103 | Regulatory apparatus of Calcium Dependent protein kinase from Arabidopsis thaliana |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)