













| General Information: |
|
| Name(s) found: |
gi|42562972
[NCBI NR]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1647 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
chromatin assembly or disassembly
[ISS]
transcription initiation [ISS] |
| Molecular Function: |
hydrolase activity, acting on ester bonds
[IEA]
transcription elongation regulator activity [IEA] RNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MARNAISDDE EDHELEDDDG EPVHGDPAEH DENDDEEDDD DVGNEYENDG FIVNDEDEEE 60
61 EEEEDEERKD SDEERQKKKK KRKKKDEGLD EDDYLLLQDN NVKFKKRQYK RLKKAQREQG 120
121 NGQGESSDDE FDSRGGTRRS AEDKIKDRLF DDVDVDDPPD DVGDEEDLVV EEDVVGSEDE 180
181 MADFIVDEDD EHGPPKRGNS KKKKYRQGSD ITAMRDANEI FGDVDELLTI RKKGLASNQR 240
241 MERRLEDEFE PTVLSEKYMT GNDDEIRQLD IPERMQISEE STGSPPVDEI SIEEESNWIY 300
301 AQLASQLRES DGTFDGRGFS VNKDDIAKFL ELHHVQKLEI PFIAMYRKEQ CRSLLDTGDF 360
361 DGANQGKKPE TKWHKVFWMI HDLDKKWLLL RKRKMALHGY YTKRYEEESR RVYDETRLNL 420
421 NQYLFESVIK SLKVAETERE VDDVDSKFNL HFPPGEIGVD EGQYKRPKRK SQYSICSKAG 480
481 LWEVANKFGY SAEQLGLALS LEKLVDELED AKETPEEMAK NFVCAMFENS LAVLKGARHM 540
541 AAVEISCEPS VKKYVRGIYM ENAVVSTSPT ADGNTVIDSF HQFSGIKWLR EKPLSKFEGA 600
601 QWLLIQKGEE EKLLQVTFKL PENYMNRLIS DCNEHYLSVG VSKYAQLWNE QRKLILEDAL 660
661 HAFLLPSMEK EARSLLTSRA KSRLLSEYGQ ALWNKVSAGP YQKKEMDINL DEEAAPRVMA 720
721 CCWGPGKPPN TFVMLDSSGE VLDVLYAGSL TSRSQNVNDQ QRKKSDQDRV LKFMMDHQPH 780
781 VVALGAVNLS CTRLKDDIYE VIFQMVEEKP RDVGHGMDDL SIVYVDESLP RLYENSRISG 840
841 EQLPQQSGNV RRAVALGRYL QNPLAMVATL CGPGREILSW KLHPLENFLQ LDEKYGMVEQ 900
901 VMVDITNQVG IDINLAASHD WFFSPLQFIS GLGPRKAASL QRSLVRAGSI FVRKDLIMHG 960
961 LGKKVFVNAA GFLRIRRSGL AASSSQFIDL LDDTRIHPES YSLAQELAKD IYDEDVRGDS1020
1021 NDDEDAIEMA IEHVRDRPAS LRKVVLDEYL ASKKRENKKE TYSNIIRELS CGFQDWRIPF1080
1081 KEPSPDEEFY MISGETEDTI AEGRIVQASV RRLQNGRAIC VLDSGLTGML MKEDFSDDGR1140
1141 DIVDLADQLK EGDILTCKIK SIQKQRYQVF LICKESEMRN NRHQHNQNVD AYYHEDRNSL1200
1201 QLVKEKARKE KELVRKHFKS RMIVHPRFQN ITADQATEYL SDKDFGESIV RPSSRGLNFL1260
1261 TLTLKIYDGV YAHKEIAEGG KENKDITSLQ CIGKTLTIGE DTFEDLDEVM DRYVDPLVSH1320
1321 LKTMLNYRKF RKGTKSEVDD LLRIEKGENP SRIVYCFGIS HEHPGTFILS YIRSTNPHHE1380
1381 YIGLYPKGFK FRKRMFEDID RLVAYFQRHI DDPLQESAPS IRSIAAKVPM RSPADHGSSG1440
1441 GSGWGSSQSE GGWKGNSDRS GSGRGGEYRN GGGRDGHPSG APRPYGGRGR GRGRGRRDDM1500
1501 NSDRQDGNGD WGNNDTGTAD GGWGNSGGGG WGSESAGKKT GGGSTGGWGS ESGGNKSDGA1560
1561 GSWGSGSGGG GSGGWGNDSG GKKSSEDGGF GSGSGGGGSD WGNESGGKKS SADGGWGSES1620
1621 GGKKSDGEGG WGNEPSSRKS DGGGGGW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
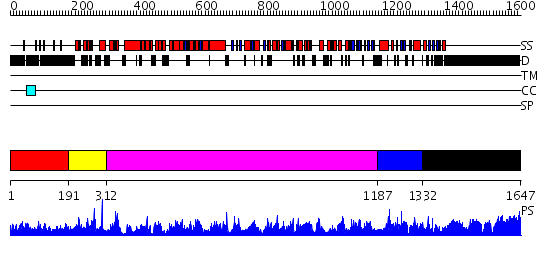
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] | 2.038996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [191..311] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [312..1186] | 1000.0 | No description for 2oceA was found. | |
| 4 | View Details | [1187..1331] | 1.7 | No description for 2bbuA was found. | |
| 5 | View Details | [1332..1647] | 51.0 | No description for 1y0fB was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)