













| General Information: |
|
| Name(s) found: |
gi|30685678
[NCBI NR]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 577 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
DNA repair
[IEA]
|
| Molecular Function: |
DNA binding
[IEA]
catalytic activity [IEA] nuclease activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGIKDLLRFM KPYILPIHIQ KYAGKRVGID AYSWLHKGAY SCSMELCLDT DGKKKLRYID 60
61 YFMHRVNLLQ HYEIIPVVVL DGGNMPCKAA TGDERHRQRK ANFDAAMVKL KEGNVAAATE 120
121 LFQRAVSVTS SMAHQLIQVL KSENVEFIVA PYEADAQLAY LSSLELEQGG IAAVITEDSD 180
181 LLAYGCKAVI FKMDRYGKGE ELVLDNVFQA VDQKPSFQNF DQELFTAMCV LAGCDFLPSV 240
241 PGVGISRAHA FISKYQSVEL VLSFLKTKKG KLVPDDYSSS FTEAVSVFQH ARVYDFDAKK 300
301 LKHLKPLSHN LLNLPVEQLE FLGPDLSPSV AVAIAEGNVD PITMKAFNHF SVSRKPLKTP 360
361 VRSFKEQEKG SSFLVCSLSK SEEIIELKRT ADEAMIDPEA IVKKKMYSKQ DSDLYKLLAQ 420
421 PNRDQVVTRP SNPSLIPDNN PFKIRKTDEI NLEIAEYGVQ ELADSFVTKS KAVDVASSSP 480
481 NSHEHEDQKE CVTIDLSKLD AAGIKQDSEK NRVKETFETT EVDLAEVQDH VNMTTKRVRG 540
541 TKPRTENFKV KTSCKSSENN KTIIKKKHSI LDFFQRL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
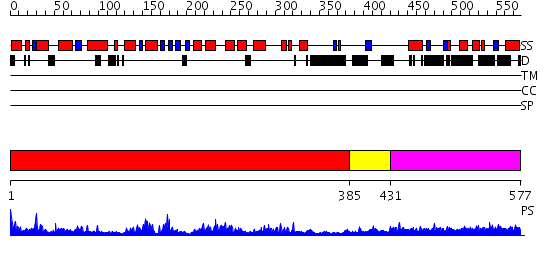
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..384] | 74.39794 | Crystal structure of the human FEN1-PCNA complex | |
| 2 | View Details | [385..430] | 4.522879 | STRUCTURE OF LARGE FRAGMENT OF ESCHERICHIA COLI DNA POLYMERASE I COMPLEXED WITH D/TMP | |
| 3 | View Details | [431..577] | 3.165997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)