













| General Information: |
|
| Name(s) found: |
G3PP2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 420 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plastid
[IDA][ISS]
|
| Biological Process: |
carbohydrate metabolic process
[IMP]
primary root development [IMP] glycolysis [ISS] |
| Molecular Function: |
NAD or NADH binding
[IEA]
catalytic activity [IEA] binding [IEA] glyceraldehyde-3-phosphate dehydrogenase activity [IEA][ISS] glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALSSLLRSA ATSAAAPRVE LYPSSSYNHS QVTSSLGFSH SLTSSRFSGA AVSTGKYNAK 60
61 RVQPIKATAT EAPPAVHRSR SSGKTKVGIN GFGRIGRLVL RIATFRDDIE VVAVNDPFID 120
121 AKYMAYMFKY DSTHGNYKGT INVIDDSTLE INGKQVKVVS KRDPAEIPWA DLGAEYVVES 180
181 SGVFTTVGQA SSHLKGGAKK VIISAPSADA PMFVVGVNEK TYLPNMDIVS NASCTTNCLA 240
241 PLAKVVHEEF GILEGLMTTV HATTATQKTV DGPSMKDWRG GRGASQNIIP SSTGAAKAVG 300
301 KVLPELNGKL TGMAFRVPTP NVSVVDLTCR LEKDASYEDV KAAIKFASEG PLRGILGYTE 360
361 EDVVSNDFLG DSRSSIFDAN AGIGLSKSFM KLVSWYDNEW GYSNRVLDLI EHMALVAASR 420
421 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
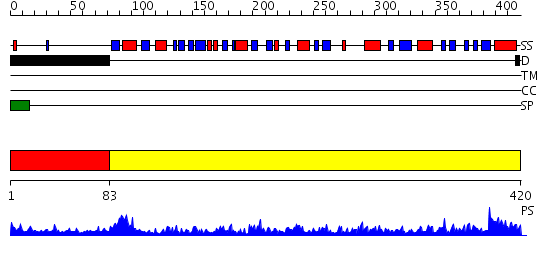
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [83..420] | 137.0 | Crystal structure of the rabbit muscle glyceraldehyde-3-phosphate dehydrogenase (GAPDH) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|