













| General Information: |
|
| Name(s) found: |
VPS33 /
YLR396C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 691 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fungal-type vacuole membrane
[IDA cytosol [IDA HOPS complex [IPI |
| Biological Process: |
late endosome to vacuole transport
[IGI vacuole fusion, non-autophagic [IDA Golgi to endosome transport [IGI |
| Molecular Function: |
ATP binding
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNRFWNTKKF SLTNADGLCA TLNEISQNDE VLVVQPSVLP VLNSLLTFQD LTQSTPVRKI 60
61 TLLDDQLSDD LPSALGSVPQ MDLIFLIDVR TSLRLPPQLL DAAQKHNLSS LHIIYCRWKP 120
121 SFQNTLEDTE QWQKDGFDLN SKKTHFPNVI ESQLKELSNE YTLYPWDLLP FPQIDENVLL 180
181 THSLYNMENV NMYYPNLRSL QSATESILVD DMVNSLQSLI FETNSIITNV VSIGNLSKRC 240
241 SHLLKKRIDE HQTENDLFIK GTLYGERTNC GLEMDLIILE RNTDPITPLL TQLTYAGILD 300
301 DLYEFNSGIK IKEKDMNFNY KEDKIWNDLK FLNFGSIGPQ LNKLAKELQT QYDTRHKAES 360
361 VHEIKEFVDS LGSLQQRQAF LKNHTTLSSD VLKVVETEEY GSFNKILELE LEILMGNTLN 420
421 NDIEDIILEL QYQYEVDQKK ILRLICLLSL CKNSLREKDY EYLRTFMIDS WGIEKCFQLE 480
481 SLAELGFFTS KTGKTDLHIT TSKSTRLQKE YRYISQWFNT VPIEDEHAAD KITNENDDFS 540
541 EATFAYSGVV PLTMRLVQML YDRSILFHNY SSQQPFILSR EPRVSQTEDL IEQLYGDSHA 600
601 IEESIWVPGT ITKKINASIK SNNRRSIDGS NGTFHAAEDI ALVVFLGGVT MGEIAIMKHL 660
661 QKILGKKGIN KRFIIIADGL INGTRIMNSI S |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
PEP3 PEP5 VAM6 VPS16 VPS33 VPS41 VPS8 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
PEP3 PEP5 VPS16 VPS33 |
| View Details | Gavin AC, et al. (2006) |
|
PEP3 PEP5 VAM6 VPS16 VPS33 VPS41 VPS8 |
| View Details | Gavin AC, et al. (2002) |

|
ATP8 LUC7 PEP3 PEP5 SNU71 VAM6 VPS16 VPS33 VPS41 VPS8 |
| View Details | Ho Y, et al. (2002) |
|
FUM1 TDA1 VPS33 |
| View Details | Qiu et al. (2008) |

|
PEP3 VPS16 VPS33 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
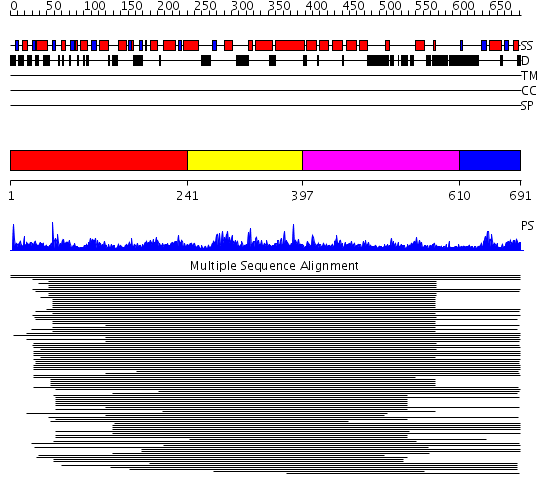
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..240] | 166.9897 | Neuronal Sec1, NSec1 | |
| 2 | View Details | [241..396] | 166.9897 | Neuronal Sec1, NSec1 | |
| 3 | View Details | [397..609] | 166.9897 | Neuronal Sec1, NSec1 | |
| 4 | View Details | [610..691] | 1.036988 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.62 |
Source: Reynolds et al. (2008)