













| General Information: |
|
| Name(s) found: |
STT4 /
YLR305C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1900 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA plasma membrane [IDA |
| Biological Process: |
MAPKKK cascade
[IDA phosphatidylethanolamine biosynthetic process [IMP vacuole organization [IMP phosphoinositide phosphorylation [IMP inositol lipid-mediated signaling [TAS [NOT]protein secretion [IMP actin cytoskeleton organization [IMP |
| Molecular Function: |
1-phosphatidylinositol 4-kinase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRFTRGLKAS SSLRAKAIGR LTKLSTGAPN DQNSNGTTLD LITHTLPIFY STNTSKIYTI 60
61 PLTLSEWEVL TSLCVAIPTT LDLVETMLKE IIAPYFLETP RQRISDVLSS KFKLEQMRNP 120
121 IELLTFQLTK FMIQACEQYP VLYENIGGII STYFERVLKI FTIKQSGLLS LVGFINAFIQ 180
181 FPNSTELTKF TWKKLAKLVL RGSFLNEVDK ILNSSATFTN DSIVQYYDAG NELSSAYLLE 240
241 LISRLQVSLI SHLLNTSHVG ANLSEFLLNQ QYQFYKFDQE VADENDDTKC IDDFFFNVRS 300
301 NKQFFTDMCK ISLQFCSESH ILDLSTDNRA RFSFDTRAHY LQTLCLIPFI EDTESELFES 360
361 FTNVVSESID KFFLSDVVTP SLIKAIVASA SLLNFFTEKL SLTLIRMFPL LVASPHITTE 420
421 TVNDVAKIFT TGLYPLNEDA IVSTIYSMNN LLAVSEDGSP VPVLRERQLT ITSGKNIEKD 480
481 YFPLRNSSAS LDGTGALLGN TTVGQLSSHD VNSGATMTYH ASLISNCVAA TTTIASYYNT 540
541 QSITALTISI LTQKVNSMSK ELDGVILNSL ARLAPNTSLT EFSLLLKFFK SRTVIATKID 600
601 DSALLKNIIK AKCVISKELL ARHFSSDLYF MYLHDLLDSI IASGEVERLE HHRPQTEISR 660
661 VADQIATYLE PLAALLPVPG DTPLDINKDE VTTNKFRNAW FNFVIHGYHL GGPIVKRNFS 720
721 FLLTIAYNSP PLASEFPANN KELSLEMNTI LRRGSSNENI KQQKQQITEY FNTNIVQYRT 780
781 TSSSKIMFLA AAVLLETIRC EAGDCSKTLL YFSDPSILSG SIEKCIAVLS VSMIRKYARL 840
841 IQKGNDAIFN SKMIAQQLNN LLLCLSHREP TLQDAAFHAC EIFIRSIPSS LCHHLSLYTL 900
901 LDMLTALFDS ILDSEAHKFE PRYEFKLKHS KTTILVPSSS SWRATTLSRL HKSAKEWVRI 960
961 LLNRSNQDTK ILLQSYISDL GEYSRLNSVE FGVSFAMDMA GLILPADKEL SRLTYYGPEK1020
1021 PNTISGFISL HSWRSKYLFD TAITSSPEDI KRQIGISTQN IRKNLTLGNK IITKDVTDFL1080
1081 DMATALLILG NGAPASLIYD IVHIPFEVFT SASLKIATNV WLTIITEKPE VAHLLLVEVC1140
1141 YCWMRSIDDN IGLYSRDHDL KGEEYQKMEY SPYDKAGINR DAKNASQAMQ PHLHVIKFFA1200
1201 SHFEGTLFQS DFLLKIFTKC ALYGIKNLYK ASLHPFARMI RHELLLFATL VLNASYKQGS1260
1261 KYMGRLSQEI TNGALSWFKR PVAWPFGSNE LKIKADLSVT RDLFLQLNKL SSLMSRHCGK1320
1321 DYKILNYFLA SEIQQIQTWL TPTEKIEGAD SNELTSDIVE ATFAKDPTLA INLLQRCYSK1380
1381 KAEDVLVGLV AKHALMCVGS PSALDLFIKG SHLSSKKDLH ATLYWAPVSP LKSINLFLPE1440
1441 WQGNSFILQF SIYSLESQDV NLAFFYVPQI VQCLRYDKTG YVERLILDTA KISVLFSHQI1500
1501 IWNMLANCYK DDEGIQEDEI KPTLDRIRER MVSSFSQSHR DFYEREFEFF DEVTGISGKL1560
1561 KPYIKKSKAE KKHKIDEEMS KIEVKPDVYL PSNPDGVVID IDRKSGKPLQ SHAKAPFMAT1620
1621 FKIKKDVKDP LTGKNKEVEK WQAAIFKVGD DCRQDVLALQ LISLFRTIWS SIGLDVYVFP1680
1681 YRVTATAPGC GVIDVLPNSV SRDMLGREAV NGLYEYFTSK FGNESTIEFQ NARNNFVKSL1740
1741 AGYSVISYLL QFKDRHNGNI MYDDQGHCLH IDFGFIFDIV PGGIKFEAVP FKLTKEMVKV1800
1801 MGGSPQTPAY LDFEELCIKA YLAARPHVEA IIECVNPMLG SGLPCFKGHK TIRNLRARFQ1860
1861 PQKTDHEAAL YMKALIRKSY ESIFTKGYDE FQRLTNGIPY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Hazbun TR, et al. (2003) |

|
STT4 YPP1 |
| View Details | Krogan NJ, et al. (2006) |

|
ATG33 BUB1 CUL3 GUT1 ISM1 MEC3 MLC1 MYO2 MYO4 RGA1 RSB1 SAT4 SHE2 SHE3 SHE4 SRC1 SSH4 STT4 TOF2 TVP18 YDR186C YOL098C |
| View Details | Gavin AC, et al. (2002) |
|
FKS1 NUM1 OPI1 OSH2 PIL1 RPN10 SCS2 STT4 SWH1 YAR044W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample zds1 from august 2005 | McCusker D, et al (2007) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
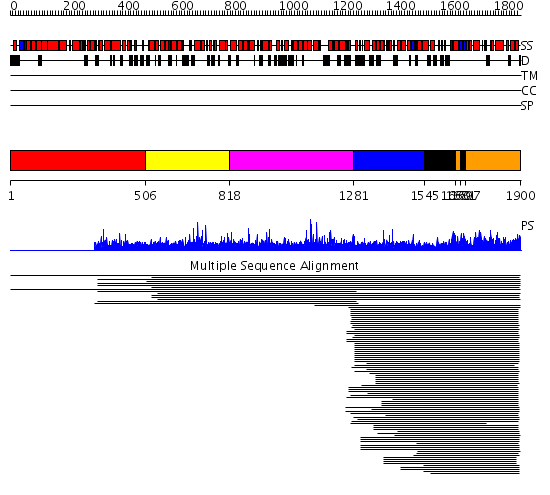
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..505] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [506..817] | 1.010931 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [818..1280] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [1281..1544] | 451.9794 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain | |
| 5 | View Details | [1545..1659] [1681..1696] |
451.9794 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain | |
| 6 | View Details | [1660..1680] [1697..1900] |
451.9794 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain | |
| 7 | View Details | [1281..1900] | 145.0 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.72 |
Source: Reynolds et al. (2008)