













| General Information: |
|
| Name(s) found: |
CDC8 /
YJR057W
[SGD]
|
| Description(s) found:
Found 58 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 216 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
mutagenesis
[IMP DNA repair [IMP DNA-dependent DNA replication [IDA dTDP biosynthetic process [IDA plasmid maintenance [IMP dUDP biosynthetic process [IDA dTTP biosynthetic process [IDA |
| Molecular Function: |
thymidylate kinase activity
[IDA uridylate kinase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMGRGKLILI EGLDRTGKTT QCNILYKKLQ PNCKLLKFPE RSTRIGGLIN EYLTDDSFQL 60
61 SDQAIHLLFS ANRWEIVDKI KKDLLEGKNI VMDRYVYSGV AYSAAKGTNG MDLDWCLQPD 120
121 VGLLKPDLTL FLSTQDVDNN AEKSGFGDER YETVKFQEKV KQTFMKLLDK EIRKGDESIT 180
181 IVDVTNKGIQ EVEALIWQIV EPVLSTHIDH DKFSFF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
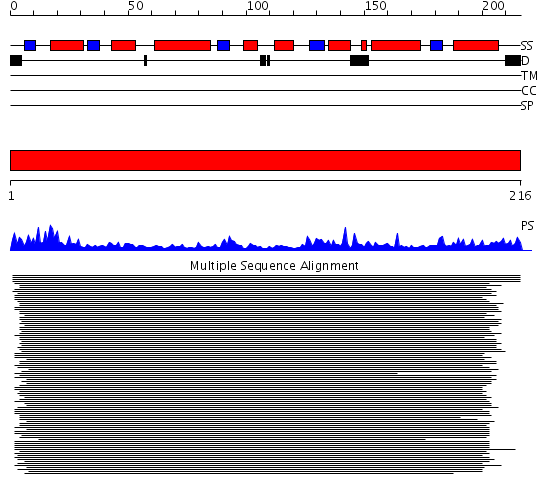
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..216] | 1257.0 | Thymidylate kinase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)