













| General Information: |
|
| Name(s) found: |
NUP157 /
YER105C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1391 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear pore
[IDA |
| Biological Process: |
mRNA export from nucleus
[IMP rRNA export from nucleus [TAS nuclear pore organization [TAS snRNP protein import into nucleus [TAS mRNA-binding (hnRNP) protein import into nucleus [TAS snRNA export from nucleus [TAS tRNA export from nucleus [TAS NLS-bearing substrate import into nucleus [TAS protein export from nucleus [TAS ribosomal protein import into nucleus [TAS |
| Molecular Function: |
structural molecule activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYSTPLKKRI DYDRETFTAS ASLGGNRLRN RPRDDQNNGK PNLSSRSFLS ERKTRKDVLN 60
61 KYGEAGNTIE SELRDVTTHV KISGLTSSEP LQLASEFVQD LSFRDRNTPI LDNPDYYSKG 120
121 LDYNFSDEVG GLGAFTPFQR QQVTNIPDEV LSQVSNTEIK SDMGIFLELN YCWITSDNKL 180
181 ILWNINNSSE YHCIDEIEHT ILKVKLVKPS PNTFVSSVEN LLIVATLFDI YILTISFNDR 240
241 THELNIFNTG LKVNVTGFNV SNIISYERTG QIFFTGATDG VNVWELQYNC SENLFNSKSN 300
301 KICLTKSNLA NLLPTKLIPS IPGGKLIQKV LEGDAGTEEE TISQLEVDQS RGVLHTLSTK 360
361 SIVRSYLITS NGLVGPVLID AAHIRRGMNA LGVKNSPLLS NRAFKIAKIV SISMCENNDL 420
421 FLAVITTTGV RLYFKGSISR RSIGSLKLDS VKFPPTSISS SLEQNKSFII GHHPLNTHDT 480
481 GPLSTQKASS TYINTTCAST IISPGIYFTC VRKRANSGEL SKGITNKALL ENKEEHKLYV 540
541 SAPDYGILKN YGKYVENTAL LDTTDEIKEI VPLTRSFNYT STPQGYANVF ASQYSAEPLK 600
601 VAVLTSNALE IYCYRTPDEV FESLIENPLP FIHSYGLSEA CSTALYLACK FNKSEHIKSS 660
661 ALAFFSAGIP GVVEIKPKSS RESGSVPPIS QNLFDKSGEC DGIVLSPRFY GSALLITRLF 720
721 SQIWEERVFV FKRASKTEKM DAFGISITRP QVEYYLSSIS VLADFFNIHR PSFVSFVPPK 780
781 GSNAITASDA ESIAMNALIL LINSIKDALS LINVFYEDID AFKSLLNTLM GAGGVYDSKT 840
841 REYFFDLKFH DLFTPNAKTK QLIKEILIEV VNANIASGTS ADYIVNVLKE RFGSFCHSAD 900
901 ILCYRAGEHL EAAQKFEMID SKISRNHLDT AIDLYERCAE NIELCELRRV VDIMVKLNYQ 960
961 PKTVGFLLRF ADKIDKGNQA QEYVSRGCNT ADPRKVFYDK RINVYTLIFE IVKSVDDYTS1020
1021 IEQSPSIANI SIFSPASSLK KRVYSVIMNS NNRFFHYCFY DWLVANKRQD YLLRLDSQFV1080
1081 LPYLKERAEK SLEISNLLWF YLFKEEHFLE AADVLYALAS SDFDLKLSER IECLARANGL1140
1141 CDSSTSFDQK PALVQLSENI HELFDIASIQ DDLLNLVRNE TRIDEDYRKQ LTLKLNGRVL1200
1201 PLSDLFNDCA DPLDYYEIKL RIFKVSQFKD EKVIQGEWNR LLDSMKNAPS PDVGSVGQES1260
1261 FLSSISNTLI RIGKTTRDTD VVFPVHFLMN KILESFIDKS SAADGSVCSM FLLAGVSHLK1320
1321 LYYILSRIIE NSEGNVELAK KEMVWLIKDW YQSDSDLRGS IAPEQIKKLE KYDPNTDPVQ1380
1381 DYVKDRHHGL K |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
GLE2 NOP14 NUP1 NUP100 NUP116 NUP133 NUP157 NUP159 NUP170 NUP188 NUP2 NUP42 POM152 |
| View Details | Qiu et al. (2008) |

|
ASM4 DBP5 GLE2 KAP123 MEX67 NDC1 NIC96 NSP1 NUP1 NUP100 NUP116 NUP120 NUP133 NUP145 NUP157 NUP159 NUP170 NUP188 NUP192 NUP2 NUP42 NUP49 NUP53 NUP57 NUP82 NUP84 NUP85 POM152 SEC13 SUS1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
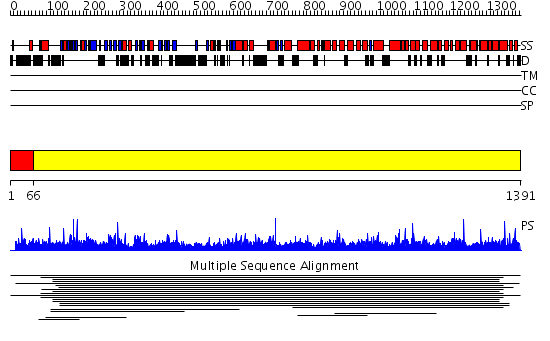
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..65] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [66..1391] | 1000.0 | Non-repetitive/WGA-negative nucleoporin No confident structure predictions are available. | |
| 3 | View Details | [258..331] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [332..437] | 1.048991 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [438..672] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [673..856] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [857..915] | 1.039993 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [916..1059] | 1.042967 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1060..1247] | 1.042993 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1248..1391] | 2.042984 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)