













| General Information: |
|
| Name(s) found: |
KRE2 /
YDR483W
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 442 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Golgi apparatus
[ISS |
| Biological Process: |
cell wall mannoprotein biosynthetic process
[IMP protein amino acid O-linked glycosylation [IMP N-glycan processing [IMP |
| Molecular Function: |
alpha-1,2-mannosyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALFLSKRLL RFTVIAGAVI VLLLTLNSNS RTQQYIPSSI SAAFDFTSGS ISPEQQVISE 60
61 ENDAKKLEQS ALNSEASEDS EAMDEESKAL KAAAEKADAP IDTKTTMDYI TPSFANKAGK 120
121 PKACYVTLVR NKELKGLLSS IKYVENKINK KFPYPWVFLN DEPFTEEFKE AVTKAVSSEV 180
181 KFGILPKEHW SYPEWINQTK AAEIRADAAT KYIYGGSESY RHMCRYQSGF FWRHELLEEY 240
241 DWYWRVEPDI KLYCDINYDV FKWMQENEKV YGFTVSIHEY EVTIPTLWQT SMDFIKKNPE 300
301 YLDENNLMSF LSNDNGKTYN LCHFWSNFEI ANLNLWRSPA YREYFDTLDH QGGFFYERWG 360
361 DAPVHSIAAA LFLPKDKIHY FSDIGYHHPP YDNCPLDKEV YNSNNCECDQ GNDFTFQGYS 420
421 CGKEYYDAQG LVKPKNWKKF RE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |

|
DOS2 KRE2 |
| View Details | Qiu et al. (2008) |
|
KRE2 KTR1 KTR4 MNN1 MNN5 OCH1 VAN1 YUR1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | De Craene, J., et al. (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
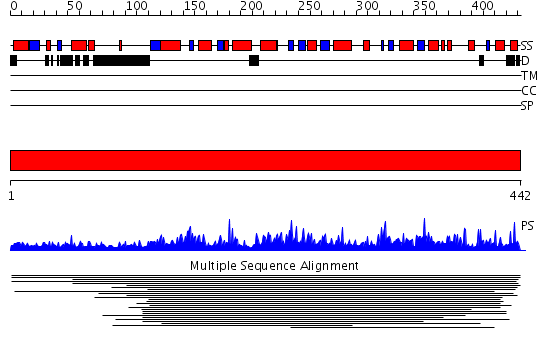
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..442] | 241.721246 | Glycolipid 2-alpha-mannosyltransferase No confident structure predictions are available. | |
| 2 | View Details | [93..442] | 169.0 | Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|