













| General Information: |
|
| Name(s) found: |
THI3 /
YDL080C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 609 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
positive regulation of transcription from RNA polymerase II promoter
[IMP thiamin biosynthetic process [IMP |
| Molecular Function: |
transcription activator activity
[IMP carboxy-lyase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSSYTQRYA LPKCIAISDY LFHRLNQLNI HTIFGLSGEF SMPLLDKLYN IPNLRWAGNS 60
61 NELNAAYAAD GYSRLKGLGC LITTFGVGEL SAINGVAGSY AEHVGILHIV GMPPTSAQTK 120
121 QLLLHHTLGN GDFTVFHRIA SDVACYTTLI IDSELCADEV DKCIKKAWIE QRPVYMGMPV 180
181 NQVNLPIESA RLNTPLDLQL HKNDPDVEKE VISRILSFIY KSQNPAIIVD ACTSRQNLIE 240
241 ETKELCNRLK FPVFVTPMGK GTVNETDPQF GGVFTGSISA PEVREVVDFA DFIIVIGCML 300
301 SEFSTSTFHF QYKTKNCALL YSTSVKLKNA TYPDLSIKLL LQKILANLDE SKLSYQPSEQ 360
361 PSMMVPRPYP AGNVLLRQEW VWNEISHWFQ PGDIIITETG ASAFGVNQTR FPVNTLGISQ 420
421 ALWGSVGYTM GACLGAEFAV QEINKDKFPA TKHRVILFMG DGAFQLTVQE LSTIVKWGLT 480
481 PYIFVMNNQG YSVDRFLHHR SDASYYDIQP WNYLGLLRVF GCTNYETKKI ITVGEFRSMI 540
541 SDPNFATNDK IRMIEIMLPP RDVPQALLDR WVVEKEQSKQ VQEENENSSA VNTPTPEFQP 600
601 LLKKNQVGY |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #16 Mitotic-New search criteria | Keck JM, et al. (2011) | |
| View Run | #18 Mitotic Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
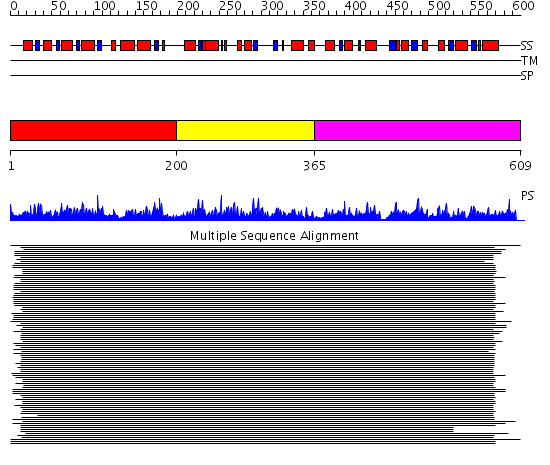
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..199] | 1921.0 | Pyruvate decarboxylase | |
| 2 | View Details | [200..364] | 1921.0 | Pyruvate decarboxylase | |
| 3 | View Details | [365..609] | 1921.0 | Pyruvate decarboxylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)