













| General Information: |
|
| Name(s) found: |
ENTF_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 1293 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQHLPLVAA QPGIWMAEKL SELPSAWSVA HYVELTGEVD SPLLARAVVA GLAQADTLRM 60
61 RFTEDNGEVW QWVDDALTFE LPEIIDLRTN IDPHGTAQAL MQADLQQDLR VDSGKPLVFH 120
121 QLIQVADNRW YWYQRYHHLL VDGFSFPAIT RQIANIYCTW LRGEPTPASP FTPFADVVEE 180
181 YQQYRESEAW QRDAAFWAEQ RRQLPPPASL SPAPLPGRSA SADILRLKLE FTDGEFRQLA 240
241 TQLSGVQRTD LALALAALWL GRLCNRMDYA AGFIFMRRLG SAALTATGPV LNVLPLGIHI 300
301 AAQETLPELA TRLAAQLKKM RRHQRYDAEQ IVRDSGRAAG DEPLFGPVLN IKVFDYQLDI 360
361 PDVQAQTHTL ATGPVNDLEL ALFPDVHGDL SIEILANKQR YDEPTLIQHA ERLKMLIAQF 420
421 AADPALLCGD VDIMLPGEYA QLAQLNATQV EIPETTLSAL VAEQAAKTPD APALADARYL 480
481 FSYREMREQV VALANLLRER GVKPGDSVAV ALPRSVFLTL ALHAIVEAGA AWLPLDTGYP 540
541 DDRLKMMLED ARPSLLITTD DQLPRFSDVP NLTSLCYNAP LTPQGSAPLQ LSQPHHTAYI 600
601 IFTSGSTGRP KGVMVGQTAI VNRLLWMQNH YPLTGEDVVA QKTPCSFDVS VWEFFWPFIA 660
661 GAKLVMAEPE AHRDPLAMQQ FFAEYGVTTT HFVPSMLAAF VASLTPQTAR QSCATLKQVF 720
721 CSGEALPADL CREWQQLTGA PLHNLYGPTE AAVDVSWYPA FGEELAQVRG SSVPIGYPVW 780
781 NTGLRILDAM MHPVPPGVAG DLYLTGIQLA QGYLGRPDLT ASRFIADPFA PGERMYRTGD 840
841 VARWLDNGAV EYLGRSDDQL KIRGQRIELG EIDRVMQALP DVEQAVTHAC VINQAAATGG 900
901 DARQLVGYLV SQSGLPLDTS ALQAQLRETL PPHMVPVVLL QLPQLPLSAN GKLDRKALPL 960
961 PELKAQAPGR APKAGSETII AAAFSSLLGC DVQDADADFF ALGGHSLLAM KLAAQLSRQV1020
1021 ARQVTPGQVM VASTVAKLAT IIDAEEDSTR RMGFETILPL REGNGPTLFC FHPASGFAWQ1080
1081 FSVLSRYLDP QWSIIGIQSP RPNGPMQTAA NLDEVCEAHL ATLLEQQPHG PYYLLGYSLG1140
1141 GTLAQGIAAR LRARGEQVAF LGLLDTWPPE TQNWQEKEAN GLDPEVLAEI NREREAFLAA1200
1201 QQGSTSTELF TTIEGNYADA VRLLTTAHSV PFDGKATLFV AERTLQEGMS PERAWSPWIA1260
1261 ELDIYRQDCA HVDIISPGTF EKIGPIIRAT LNR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
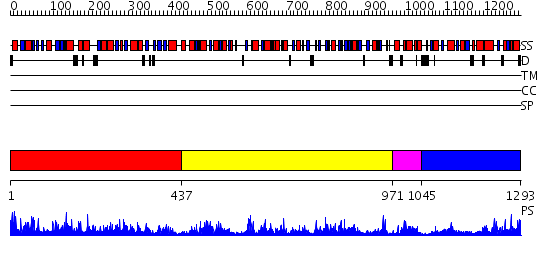
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..436] | 37.0 | VibH | |
| 2 | View Details | [437..970] | 119.0 | Phenylalanine activating domain of gramicidin synthetase 1 | |
| 3 | View Details | [971..1044] | 10.30103 | No description for 2gdwA was found. | |
| 4 | View Details | [1045..1293] | 17.0 | Surfactin synthetase, SrfA |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.49 |
Source: Reynolds et al. (2008)