













| General Information: |
|
| Name(s) found: |
LONM4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 942 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrion [IDA] |
| Biological Process: |
proteolysis
[IEA]
|
| Molecular Function: |
nucleotide binding
[IEA]
ATP binding [IEA][ISS] serine-type endopeptidase activity [IEA] serine-type peptidase activity [ISS] nucleoside-triphosphatase activity [IEA] ATP-dependent peptidase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLKFLTPTAY ASHHVTPATR FRSTPVKNLL FKQLTLLTGW NRSSYELGRR SFSSDLDSDT 60
61 KSSTTTVSAK PHLDDCLTVI ALPLPHKPLI PGFYMPIYVK DPKVLAALQE SRRQQAPYAG 120
121 AFLLKDDASS DSSSSSETEN ILEKLKGKEL INRIHEVGTL AQISSIQGEQ VILIGHRQLR 180
181 ITEMVSESED PLTVKVDHLK DKPYDKDDDV IKATYFQVMS TLRDVLKTTS LWRDHVRTYT 240
241 QACSLHIWHC LRHIGEFNYP KLADFGAGIS GANKHQNQGV LEELDVHKRL ELTLELVKKE 300
301 VEINKIQESI AKAVEEKFSG DRRRIILKEQ INAIKKELGG ETDSKSALSE KFRGRIDPIK 360
361 DKIPGHVLKV IEEELKKLQL LETSSSEFDV TCNYLDWLTV LPWGNFSDEN FNVLRAEKIL 420
421 DEDHYGLSDV KERILEFIAV GGLRGTSQGK IICLSGPTGV GKTSIGRSIA RALDRKFFRF 480
481 SVGGLSDVAE IKGHRRTYIG AMPGKMVQCL KNVGTENPLV LIDEIDKLGV RGHHGDPASA 540
541 MLELLDPEQN ANFLDHYLDV PIDLSKVLFV CTANVTDTIP GPLLDRMEVI TLSGYITDEK 600
601 MHIARDYLEK TARRDCGIKP EQVDVSDAAF LSLIEHYCRE AGVRNLQKQI EKIFRKIALK 660
661 LVRKAASTEV PRISDDVTTD TEETKSLAKT DLESPETSAE GSTVLTDELA TGDPTESTTE 720
721 QSGEVAETVE KYMIDESNLS DYVGKPVFQE EKIYEQTPVG VVMGLAWTSM GGSTLYIETT 780
781 FVEEGEGKGG LHITGRLGDV MKESAEIAHT VARRIMLEKE PENKLFANSK LHLHVPAGAT 840
841 PKDGPSAGCT MITSLLSLAL KKPVRKDLAM TGEVTLTGRI LAIGGVKEKT IAARRSQVKV 900
901 IIFPEANRRD FDELARNVKE GLEVHFVDEY EQIFELAFGY DH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
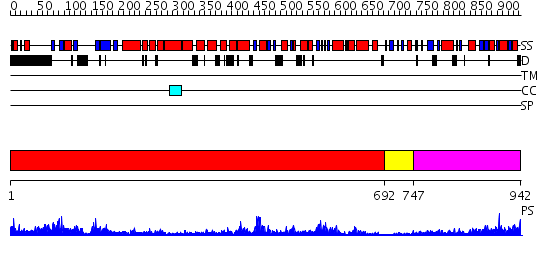
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..691] | 110.0 | Crystal Structure Analysis of ClpB | |
| 2 | View Details | [692..746] | 14.045757 | ADP Bound E. coli Clamp Loader Complex | |
| 3 | View Details | [747..942] | 32.09691 | Catalytic domain of E.coli Lon protease |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)