













| General Information: |
|
| Name(s) found: |
chn-1 /
CE29991
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 266 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ubiquitin ligase complex
[IEA cytoplasm [IDA |
| Biological Process: |
locomotion
[IGI nematode larval development [IMP protein polyubiquitination [IDA sarcomere organization [IGI oviposition [IGI protein autoubiquitination [IDA protein ubiquitination [IDA |
| Molecular Function: |
ubiquitin-protein ligase activity
[IDA ubiquitin protein ligase binding [IPI chaperone binding [IPI Hsp70 protein binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSGAEQHNT NGKKCYMNKR YDDAVDHYSK AIKVNPLPKY YQNRAMCYFQ LNNLKMTEED 60
61 CKRALELSPN EVKPLYFLGN VFLQSKKYSE AISCLSKALY HNAVITNAPD IENALKRARH 120
121 QKYEEEESKR IVQDVEFHTY LESLIEKDRQ ENSENPEELQ RADMAKKRLT ELTLATQEKR 180
181 QNREVPEMLC GKITLELMKE PVIVPSGITY DREEIVQHLR RIGHFDPVTR KPLTENEIIP 240
241 NYALKEVIEK FLDDNPWAKY EPGGMV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
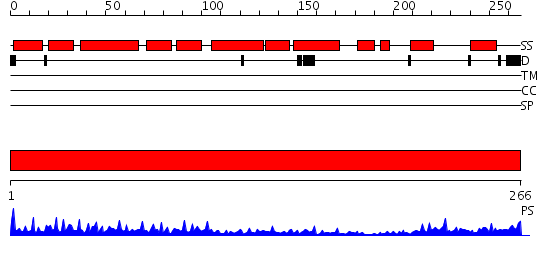
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..266] | 30.522879 | Crystal structure of the CHIP U-box E3 ubiquitin ligase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)