













| General Information: |
|
| Name(s) found: |
rnc1 /
SPCC757.09c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 398 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
negative regulation of MAPKKK cascade
[IGI mRNA stabilization [IMP negative regulation of protein kinase activity [IGI |
| Molecular Function: |
protein binding
[IPI mRNA binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAYNHFSIPK NIEEKENSFF DVTFQDEPDE TTSTATGIAK VSIPTPKPST PLSTLTNGST 60
61 IQQSMTNQPE PTSQVPPISA KPPMDDATYA TQQLTLRALL STREAGIIIG KAGKNVAELR 120
121 STTNVKAGVT KAVPNVHDRV LTISGPLENV VRAYRFIIDI FAKNSTNPDG TPSDANTPRK 180
181 LRLLIAHSLM GSIIGRNGLR IKLIQDKCSC RMIASKDMLP QSTERTVEIH GTVDNLHAAI 240
241 WEIGKCLIDD WERGAGTVFY NPVSRLTQPL PSLASTASPQ QVSPPAAPST TSGEAIPENF 300
301 VSYGAQVFPA TQMPFLQQPK VTQNISIPAD MVGCIIGRGG SKISEIRRTS GSKISIAKEP 360
361 HDETGERMFT ITGTHEENEK ALFLLYQQLE MEKDRRSH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
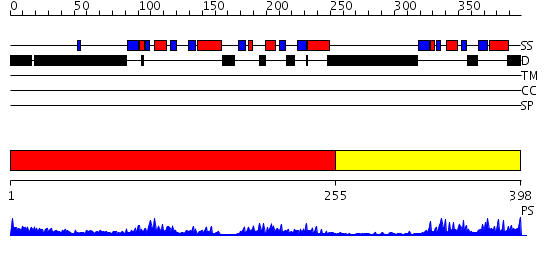
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..254] | 29.522879 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 2 | View Details | [255..398] | 15.0 | Crsytal Structure of domain 3 of human alpha polyC binding protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)