













| General Information: |
|
| Name(s) found: |
crk1 /
SPBC19F8.07
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 335 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA cytosol [IDA holo TFIIH complex [ISS |
| Biological Process: |
phosphorylation of RNA polymerase II C-terminal domain
[IMP positive regulation of phosphorylation [IC P-TEFb-cap methyltransferase complex localization [IMP regulation of gene-specific transcription elongation from RNA polymerase II promoter [IMP regulation of catalytic activity [IC regulation of cell cycle [IGI positive regulation of transcription from RNA polymerase II promoter [ISS protein amino acid phosphorylation [IC |
| Molecular Function: |
cyclin-dependent protein kinase activating kinase activity
[IGI ATP binding [IEA] RNA polymerase II carboxy-terminal domain kinase activity [IDA general RNA polymerase II transcription factor activity [ISS cyclin-dependent protein kinase activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDIEKSDKWT YVKERKVGEG TYAVVFLGRQ KETNRRVAIK KIKVGQFKDG IDISALREIK 60
61 FLRESRHDNV IELVDVFSTK SNLNIILEFL DSDLEMLIKD KFIVFQPAHI KSWMVMLLRG 120
121 LHHIHSRFIL HRDLKPNNLL ISSDGVLKLA DFGLSRDFGT PSHMSHQVIT RWYRPPELFM 180
181 GCRSYGTGVD MWSVGCIFAE LMLRTPYLPG ESDLDQLNVI FRALGTPEPE VIKSMQQLPN 240
241 YVEMKHIPPP NGGMEALFSA AGHEEIDLLK MMLDYNPYRR PTAQQALEHH YFSALPKPTH 300
301 PSLLPRKGGE EGIKHVSSDL QRQNNFPMRA NIKFV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
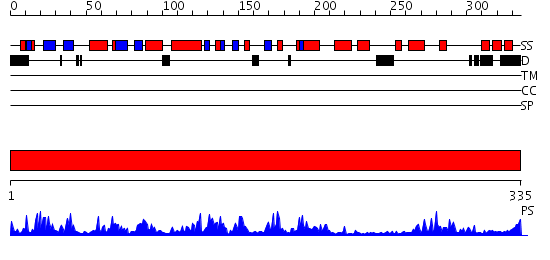
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..335] | 82.045757 | Crystal Structure of Human CDK7 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)