













| General Information: |
|
| Name(s) found: |
spn-B-PA /
FBpp0082392
[FlyBase]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 341 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
DNA repair
[ISS][NAS]
intracellular mRNA localization [IMP][TAS] reciprocal meiotic recombination [TAS] germarium-derived oocyte fate determination [IGI] chromosome condensation [TAS] DNA recombination [ISS] polarity specification of dorsal/ventral axis [IMP] oocyte differentiation [NAS] DNA metabolic process [IEA] meiosis [TAS] double-strand break repair [TAS] polarity specification of anterior/posterior axis [IMP] regulation of translation [TAS] oogenesis [IGI][TAS] karyosome formation [IMP] |
| Molecular Function: |
DNA binding
[IEA]
ATP binding [IEA] DNA-dependent ATPase activity [IEA] recombinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTNFLDQLPK HIREIAEQVN VLAPEEVLTT PKVRFLDTRQ QSLHTIVRKC TPDDVRVLKD 60
61 AAAKWLAEMP QSADSLFKPL VNVRWSRVSF GCSALDRCTG GGVVTRGITE LCGAAGVGKT 120
121 QLLLQLSLCV QLPRELGGLG KGVAYICTES SFPARRLLQM SKACEKRHPE MELNFLGNIF 180
181 VENHIEAEPL LACVINRIPR LMQQHGIGLI IIDSVAAIFR LYNDYLERAR HMRRLADALL 240
241 SYADKYNCAV VCVNQVATRD GQDEIPCLGL QWAHLGRTRL RVSRVPKQHR MGDQLITVRK 300
301 LEILYSPETP NDFAEFLITA EGVVNVPEPS VPSPPAKMRR L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
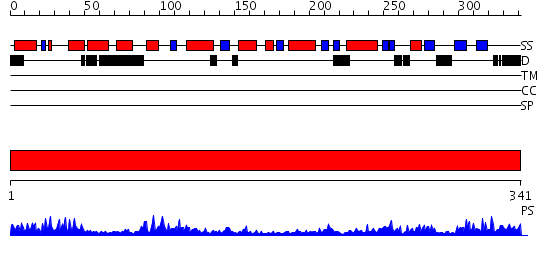
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..341] | 65.69897 | Crystal structure of the human Dmc1 protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.66 |
Source: Reynolds et al. (2008)