













| General Information: |
|
| Name(s) found: |
PLP2 /
YOR281C
[SGD]
|
| Description(s) found:
Found 40 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 286 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IPI |
| Biological Process: |
positive regulation of transcription from RNA polymerase II promoter by pheromones
[IMP |
| Molecular Function: |
GTPase inhibitor activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQNEPMFQVQ VDESEDSEWN DILRAKGVIP ERAPSPTAKL EEALEEAIAK QHENRLEDKD 60
61 LSDLEELEDD EDEDFLEAYK IKRLNEIRKL QERSKFGEVF HINKPEYNKE VTLASQGKKY 120
121 EGAQTNDNGE EDDGGVYVFV HLSLQSKLQS RILSHLFQSA ACKFREIKFV EIPANRAIEN 180
181 YPESNCPTLI VYYRGEVIKN MITLLELGGN NSKMEDFEDF MVKVGAVAEG DNRLIMNRDD 240
241 EESREERKLH YGEKKSIRSG IRGKFNVGIG GNDDGNINDD DDGFFD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |

|
CCT2 CCT7 PLP2 VID27 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
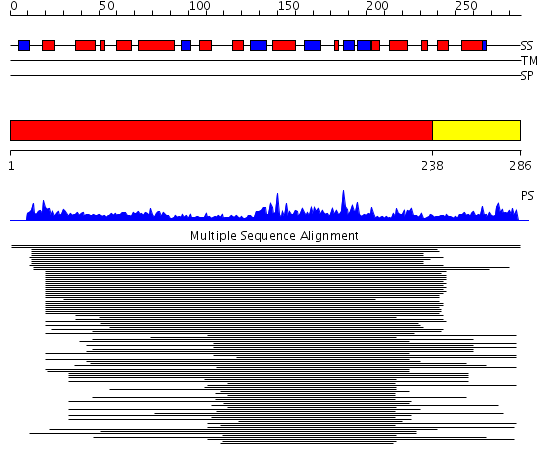
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..237] | 59.27013 | Phosducin | |
| 2 | View Details | [238..286] | 5.045757 | Protein disulfide isomerase, PDI |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)