













| General Information: |
|
| Name(s) found: |
APC1 /
YNL172W
[SGD]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1748 amino acids |
Gene Ontology: |
|
| Cellular Component: |
anaphase-promoting complex
[TAS |
| Biological Process: |
mitotic spindle elongation
[TAS cyclin catabolic process [TAS mitotic sister chromatid segregation [TAS anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process [IDA mitotic metaphase/anaphase transition [TAS protein ubiquitination [IPI |
| Molecular Function: |
ubiquitin-protein ligase activity
[TAS protein binding [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSKPSTRND YLPRETHNGE YTGDSPEWQL QINITNKIGG INGDIWLSRD GRSVKWCIED 60
61 QCLRQFTYNQ KIIKAGYIDF EKTPDCFVVV LSDIAHVYML KNGGSTTVCF PFQIGNAFWY 120
121 ANGVILERET SASFMDGGYD LKPIEFDLKH KYITLTDPMA PFGLISITNT FKGGINSASG 180
181 NKTDILQDFQ LVLFPSDKDK CIAVFLDRNS KVLRFYYSRI LSSDQSRKGE LTISSTKKTG 240
241 LDAAGNSQKS GGISKDLRKF SHLTRRSTSI NSNSHDFNAA ERVLSGNVGN ASGRTDIFAL 300
301 PSSCSRRSLS ATLDRMGNNI APTNRAAPSG FFDSSSANTA THSNITPVSQ PMQQQQQEYL 360
361 NQTATSSKDI VLTEISSLKL PDDIIFTSRR LSSILSKLKF LSLRFERREG LLIFHEPTHF 420
421 CKIWLIDLLP DVLDSIPFKI YGNSPQNMIR LENLKLKEPS RIQAMYIHEL LESCLILVSE 480
481 GQNKEEYKAC LYDPFVKITS PSKNISEELT KQNSLPSLQK LFPYPETSFT KLCFEAVKYI 540
541 TSPAFNISFI FLWQSAYSIL LSRANDDVVG GLKMEHDAFS LVLSLLILPI PSSSAQEYQE 600
601 YKEIYERDLF QHLKQDSEIT SSVLPRIVIG LHLIREEYSL NVLCRNEHAL LGQFLRFATA 660
661 AMGWPDLWQS YYVPKMDSES KTFLHPREQN STFFHPLDEP PSITKSLYSI TENSSIPLCP 720
721 FISFSRLVAT DTQVELRITP RSFKILGLYE LVHSPNFLPD YVLGILSSFK VDKDELQTYP 780
781 LGILVPLQNI LKILEDKLSE VRDNLELLDR ADLQRCSAII NSIRSDSKEV LKRGQRDSYM 840
841 LCKVPLAKNR SSLSKKPSDI YSILSEIVKS ASQVPLDGSA MRMSNIQDDE DIDEGRSLKL 900
901 NAGLIFSEDK RFTHVVSLLA YYRPTKTQFF TTKTEYAQIL AQKKYFAKIM ALRTCTNGVG 960
961 WGAVAYATEK PISTQKWVIQ PLNLISVFPD DTKITVKAPE DIAHDIVEWG QFHAGVSSGL1020
1021 RISKKATGIT GSWIAFNKPK ELDAYHGGFL LGLGLNGHLK NLEEWHIYNY LSPRNTHISI1080
1081 GLLLGMSSSM KGSMDSKLIK VISVHLVAFL PSGSSDLNID LKLQTAGIIG MGMLYLNSRH1140
1141 KRMSDSIFAQ LVSLLNVNDE MVADEEYRLA AGISLGLINL GAGQTKLRKW DSSLLGLGDD1200
1201 LPEDVYDSSD VEQNVMYEDL TTKLLEIVTS TYDVENDWIP ENSQIGAVIA IMFLFLKSNN1260
1261 FGISNMLKVD LKEILKANIN TRPELLMYRE WASNMILWEF IGDDLSFIMK DVDIGVKFSE1320
1321 LNTDLLPIYY TMAGRILAMG IRFASTGNLK IRNILLSLVD KFLPLYQYPG KQNLDFRLTI1380
1381 SVINVLTNVI VVSLSMVMCA SGDLEVLRRV KYLHEVASGP YSDLFQEIPS SKSDVSGVTQ1440
1441 VTSNTNTPGN SDRERVDETA ASLDDERSSN GSDISDPTAY LEDKKDIDDH YGKFISTNLA1500
1501 LGFLFLGSGQ YALNTSTLES IAFLSMSVLP TYTTPHPLQE LKHFWSIAVE PRCLVIKDIS1560
1561 TGDAVNNVPI ELVVEEDVEK EEVIREISTP CLLPDFSKIK SIRVKMHGYF PLEVNFTKDY1620
1621 SASDFFSGGT IIYIQRKSES VFENKASFRN VEDIHVALKR KAAESKNYSR LNLKNEQGNT1680
1681 TSSQLVESLG IQDLTMVELD TLLSAGNNTA LTDSESYNLG LLCSDKNSGD ILDCQLELWY1740
1741 KSFGPHDE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
APC1 APC2 APC4 APC5 CDC16 CDC23 CDC27 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
APC1 APC11 APC2 APC4 APC5 APC9 CDC16 CDC23 CDC26 CDC27 DOC1 |
| View Details | Krogan NJ, et al. (2006) |

|
APC1 APC2 APC4 APC5 CDC16 CDC23 CDC26 CDC27 DOC1 FMP52 MND2 SSP2 |
| View Details | Gavin AC, et al. (2006) |

|
APC1 APC2 APC4 APC5 APC9 CDC16 CDC23 CDC27 |
| View Details | Gavin AC, et al. (2002) |

|
ACT1 APC1 APC2 CDC16 CDC23 CDC27 |
| View Details | Ho Y, et al. (2002) |
|
APC1 TMA29 YCK1 |
| View Details | Qiu et al. (2008) |

|
APC1 APC11 APC2 APC4 APC5 APC9 BUL2 CDC16 CDC23 CDC26 CDC27 DOC1 HRT1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
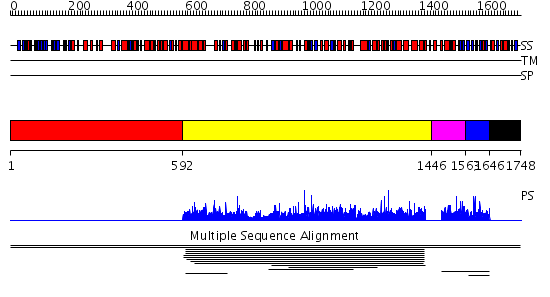
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..591] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [592..1445] | 5.009001 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [1446..1562] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [1563..1645] | 1.003977 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [1646..1748] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [885..970] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [971..1025] | 1.042991 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1026..1748] | 0.951 | No description for 2ie3A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 8 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.63 |
Source: Reynolds et al. (2008)