













| General Information: |
|
| Name(s) found: |
GAS2 /
YLR343W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 555 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
ascospore wall assembly
[IGI |
| Molecular Function: |
1,3-beta-glucanosyltransferase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNKKQNFYAA IIVAIFLCLQ LSHGSSGVSF EKTPAIKIVG NKFFDSESGE QFFIKGIAYQ 60
61 LQRSEEELSN ANGAFETSYI DALADPKICL RDIPFLKMLG VNTLRVYAID PTKSHDICME 120
121 ALSAEGMYVL LDLSEPDISI NRENPSWDVH IFERYKSVID AMSSFPNLLG YFAGNEVTND 180
181 HTNTFASPFV KAAIRDAKEY ISHSNHRKIP VGYSTNDDAM TRDNLARYFV CGDVKADFYG 240
241 INMYEWCGYS TYGTSGYRER TKEFEGYPIP VFFSEFGCNL VRPRPFTEVS ALYGNKMSSV 300
301 WSGGLAYMYF EEENEYGVVK INDNDGVDIL PDFKNLKKEF AKADPKGITE EEYLTAKEPT 360
361 EVESVECPHI AVGVWEANEK LPETPDRSKC ACLDEILPCE IVPFGAESGK YEEYFSYLCS 420
421 KVDCSDILAN GKTGEYGEFS DCSVEQKLSL QLSKLYCKIG ANDRHCPLND KNVYFNLESL 480
481 QPLTSESICK NVFDSIRNIT YNHGDYSKSN PSRSKESLNV KYPSSEEREN DGTIAFKTSG 540
541 FVILLISMIA AGILL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
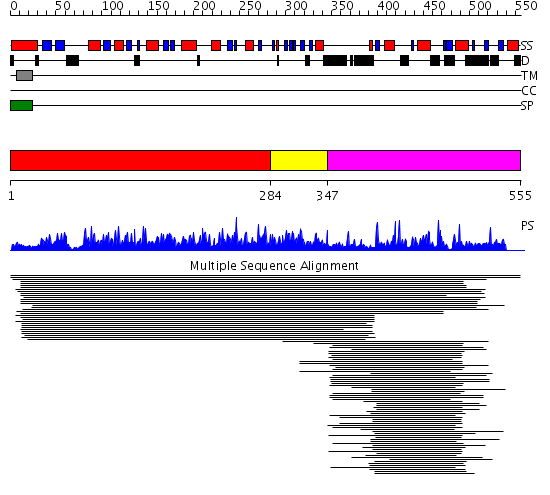
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..283] | 14.38 | Endoglucanase Cel5a | |
| 2 | View Details | [284..346] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [347..555] | 38.091987 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|