













| General Information: |
|
| Name(s) found: |
RAD5 /
YLR032W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1169 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear chromatin
[IDA |
| Biological Process: |
DNA repair
[IMP telomere maintenance [IMP double-strand break repair [IGI |
| Molecular Function: |
ATPase activity
[TAS single-stranded DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSHIEQEERK RFFNDDLDTS ETSLNFKSEN KESFLFANSH NDDDDDVVVS VSDTTEGEGD 60
61 RSIVPVRREI EEEGQNQFIT ELLRIIPEMP KDLVMELNEK FGSQEEGLSL ALSHYFDHNS 120
121 GTSISKIPSS PNQLNTLSDT SNSTLSPSSF HPKRRRIYGF RNQTRLEDKV TWKRFIGALQ 180
181 VTGMATRPTV RPLKYGSQMK LKRSSEEISA TKVYDSRGRK KASMASLVRI FDIQYDREIG 240
241 RVSEDIAQIL YPLLSSHEIS FEVTLIFCDN KRLSIGDSFI LQLDCFLTSL IFEERNDGES 300
301 LMKRRRTEGG NKREKDNGNF GRTLTETDEE LESRSKRLAL LKLFDKLRLK PILDEQKALE 360
361 KHKIELNSDP EIIDLDNDEI CSNQVTEVHN NLRDTQHEEE TMNLNQLKTF YKAAQSSESL 420
421 KSLPETEPSR DVFKLELRNY QKQGLTWMLR REQEFAKAAS DGEASETGAN MINPLWKQFK 480
481 WPNDMSWAAQ NLQQDHVNVE DGIFFYANLH SGEFSLAKPI LKTMIKGGIL SDEMGLGKTV 540
541 AAYSLVLSCP HDSDVVDKKL FDIENTAVSD NLPSTWQDNK KPYASKTTLI VVPMSLLTQW 600
601 SNEFTKANNS PDMYHEVYYG GNVSSLKTLL TKTKTPPTVV LTTYGIVQNE WTKHSKGRMT 660
661 DEDVNISSGL FSVNFYRIII DEGHNIRNRT TVTSKAVMAL QGKCKWVLTG TPIINRLDDL 720
721 YSLVKFLELD PWRQINYWKT FVSTPFESKN YKQAFDVVNA ILEPVLLRRT KQMKDKDGKP 780
781 LVELPPKEVV IKRLPFSKSQ DLLYKFLLDK AEVSVKSGIA RGDLLKKYST ILVHILRLRQ 840
841 VCCHPGLIGS QDENDEDLSK NNKLVTEQTV ELDSLMRVVS ERFDNSFSKE ELDAMIQRLK 900
901 VKYPDNKSFQ SLECSICTTE PMDLDKALFT ECGHSFCEKC LFEYIEFQNS KNLGLKCPNC 960
961 RNQIDACRLL ALVQTNSNSK NLEFKPYSPA SKSSKITALL KELQLLQDSS AGEQVVIFSQ1020
1021 FSTYLDILEK ELTHTFSKDV AKIYKFDGRL SLKERTSVLA DFAVKDYSRQ KILLLSLKAG1080
1081 GVGLNLTCAS HAYMMDPWWS PSMEDQAIDR LHRIGQTNSV KVMRFIIQDS IEEKMLRIQE1140
1141 KKRTIGEAMD TDEDERRKRR IEEIQMLFE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
ALT2 RAD5 RRP1 |
| View Details | Qiu et al. (2008) |
|
PCC1 RAD18 RAD5 RAP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
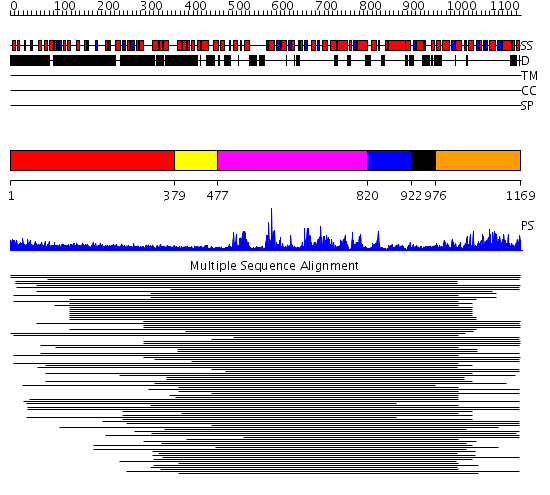
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..378] | 5.209996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [379..476] | 11.29 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 3 | View Details | [477..819] | 11.29 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 4 | View Details | [820..921] | 1.345993 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [922..975] | N/A | Confident ab initio structure predictions are available. | |
| 6 | View Details | [976..1169] | 2.522879 | Nucleotide excision repair enzyme UvrB |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)