













| General Information: |
|
| Name(s) found: |
HBS1 /
YKR084C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 611 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
translation
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAYSDYSDGA DDMPDFHDEG EFDDYLNDDE YDLMNEVFPT LKAQLQDYQG WDNLSLKLAL 60
61 FDNNFDLEST LAELKKTLKK KKTPKKPIAA ANGSANVTQK LANISISQQR PNDRLPDWLD 120
121 EEESEGERNG EEANDEKTVQ RYYKTTVPTK PKKPHDISAF VKSALPHLSF VVLGHVDAGK 180
181 STLMGRLLYD LNIVNQSQLR KLQRESETMG KSSFKFAWIM DQTNEERERG VTVSICTSHF 240
241 STHRANFTIV DAPGHRDFVP NAIMGISQAD MAILCVDCST NAFESGFDLD GQTKEHMLLA 300
301 SSLGIHNLII AMNKMDNVDW SQQRFEEIKS KLLPYLVDIG FFEDNINWVP ISGFSGEGVY 360
361 KIEYTDEVRQ WYNGPNLMST LENAAFKISK ENEGINKDDP FLFSVLEIIP SKKTSNDLAL 420
421 VSGKLESGSI QPGESLTIYP SEQSCIVDKI QVGSQQGQST NHEETDVAIK GDFVTLKLRK 480
481 AYPEDIQNGD LAASVDYSSI HSAQCFVLEL TTFDMNRPLL PGTPFILFIG VKEQPARIKR 540
541 LISFIDKGNT ASKKKIRHLG SKQRAFVEIE LIEVKRWIPL LTAHENDRLG RVVLRKDGRT 600
601 IAAGKISEIT Q |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
CAM1 EFB1 HBS1 TEF1, TEF2 TEF4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
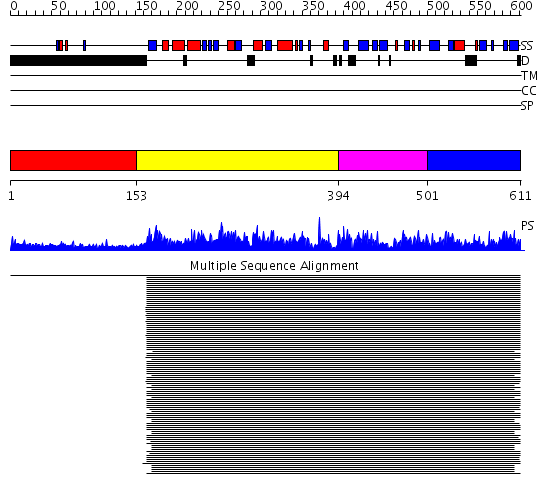
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..152] | 3.590996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [153..393] | 1580.0 | Elongation factor eEF-1alpha, domain 2; Elongation factor eEF-1alpha, C-terminal domain; Elongation factor eEF-1alpha, N-terminal (G) domain | |
| 3 | View Details | [394..500] | 1580.0 | Elongation factor eEF-1alpha, domain 2; Elongation factor eEF-1alpha, C-terminal domain; Elongation factor eEF-1alpha, N-terminal (G) domain | |
| 4 | View Details | [501..611] | 1580.0 | Elongation factor eEF-1alpha, domain 2; Elongation factor eEF-1alpha, C-terminal domain; Elongation factor eEF-1alpha, N-terminal (G) domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)