













| General Information: |
|
| Name(s) found: |
FOX2 /
YKR009C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 900 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisomal matrix
[TAS]
|
| Biological Process: |
fatty acid beta-oxidation
[TAS]
|
| Molecular Function: |
3-hydroxyacyl-CoA dehydrogenase activity
[TAS]
enoyl-CoA hydratase activity [TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPGNLSFKDR VVVITGAGGG LGKVYALAYA SRGAKVVVND LGGTLGGSGH NSKAADLVVD 60
61 EIKKAGGIAV ANYDSVNENG EKIIETAIKE FGRVDVLINN AGILRDVSFA KMTEREFASV 120
121 VDVHLTGGYK LSRAAWPYMR SQKFGRIINT ASPAGLFGNF GQANYSAAKM GLVGLAETLA 180
181 KEGAKYNINV NSIAPLARSR MTENVLPPHI LKQLGPEKIV PLVLYLTHES TKVSNSIFEL 240
241 AAGFFGQLRW ERSSGQIFNP DPKTYTPEAI LNKWKEITDY RDKPFNKTQH PYQLSDYNDL 300
301 ITKAKKLPPN EQGSVKIKSL CNKVVVVTGA GGGLGKSHAI WFARYGAKVV VNDIKDPFSV 360
361 VEEINKLYGE GTAIPDSHDV VTEAPLIIQT AISKFQRVDI LVNNAGILRD KSFLKMKDEE 420
421 WFAVLKVHLF STFSLSKAVW PIFTKQKSGF IINTTSTSGI YGNFGQANYA AAKAAILGFS 480
481 KTIALEGAKR GIIVNVIAPH AETAMTKTIF SEKELSNHFD ASQVSPLVVL LASEELQKYS 540
541 GRRVIGQLFE VGGGWCGQTR WQRSSGYVSI KETIEPEEIK ENWNHITDFS RNTINPSSTE 600
601 ESSMATLQAV QKAHSSKELD DGLFKYTTKD CILYNLGLGC TSKELKYTYE NDPDFQVLPT 660
661 FAVIPFMQAT ATLAMDNLVD NFNYAMLLHG EQYFKLCTPT MPSNGTLKTL AKPLQVLDKN 720
721 GKAALVVGGF ETYDIKTKKL IAYNEGSFFI RGAHVPPEKE VRDGKRAKFA VQNFEVPHGK 780
781 VPDFEAEIST NKDQAALYRL SGDFNPLHID PTLAKAVKFP TPILHGLCTL GISAKALFEH 840
841 YGPYEELKVR FTNVVFPGDT LKVKAWKQGS VVVFQTIDTT RNVIVLDNAA VKLSQAKSKL 900
901 |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
CYK3 FOX2 |
| View Details | Qiu et al. (2008) |
|
DCI1 FOX2 MDH3 POT1 POX1 SPS19 TES1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Green EM, et al (2005) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
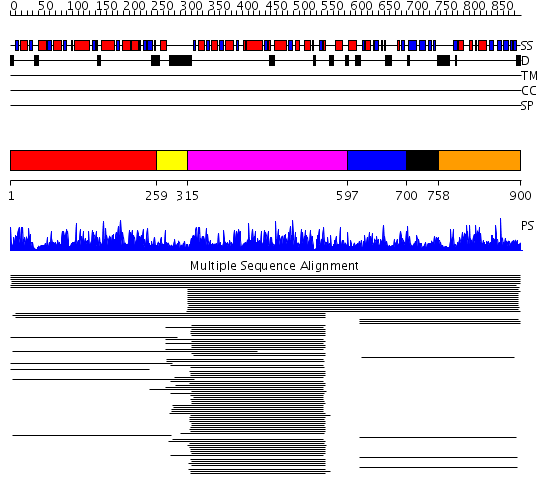
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..258] | 17.221849 | Enoyl-ACP reductase | |
| 2 | View Details | [259..314] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [315..596] | 417.228787 | beta-keto acyl carrier protein reductase | |
| 4 | View Details | [597..699] | 1.026982 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [700..757] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [758..900] | 11.039997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.65 |
Source: Reynolds et al. (2008)