













| General Information: |
|
| Name(s) found: |
APE2 /
YKL157W
[SGD]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 935 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[NAS mitochondrion [IDA cell wall-bounded periplasmic space [NAS |
| Biological Process: |
peptide metabolic process
[IMP |
| Molecular Function: |
leucyl aminopeptidase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPIVRWLLLK SAVRGSSLIG KAHPCLRSIA AHPRYLSNVY SPPAGVSRSL RINVMWKQSK 60
61 LTPPRFVKIM NRRPLFTETS HACAKCQKTS QLLNKTPNRE ILPDNVVPLH YDLTVEPDFK 120
121 TFKFEGSVKI ELKINNPAID TVTLNTVDTD IHSAKIGDVT SSEIISEEEQ QVTTFAFPKG 180
181 TMSSFKGNAF LDIKFTGILN DNMAGFYRAK YEDKLTGETK YMATTQMEPT DARRAFPCFD 240
241 EPNLKASFAI TLVSDPSLTH LSNMDVKNEY VKDGKKVTLF NTTPKMSTYL VAFIVAELKY 300
301 VESKNFRIPV RVYATPGNEK HGQFAADLTA KTLAFFEKTF GIQYPLPKMD NVAVHEFSAG 360
361 AMENWGLVTY RVVDLLLDKD NSTLDRIQRV AEVVQHELAH QWFGNLVTMD WWEGLWLNEG 420
421 FATWMSWYSC NEFQPEWKVW EQYVTDTLQH ALSLDSLRSS HPIEVPVKKA DEINQIFDAI 480
481 SYSKGASLLR MISKWLGEET FIKGVSQYLN KFKYGNAKTE DLWDALADAS GKDVRSVMNI 540
541 WTKKVGFPVI SVSEDGNGKI TFRQNRYLST ADVKPDEDKT IYPVFLALKT KNGVDSSVVL 600
601 SERSKTIELE DPTFFKVNSE QSGIYITSYT DERWAKLGQQ ADLLSVEDRV GLVADVKTLS 660
661 ASGYTSTTNF LNLVSKWNNE KSFVVWDQII NSISSMKSTW LFEPKETQDA LDNFTKQLIS 720
721 GMTHHLGWEF KSSDSFSTQR LKVTMFGAAC AARDADVEKA ALKMFTDYCS GNKEAIPALI 780
781 KPIVFNTVAR VGGAENYEKV YKIYLDPISN DEKLAALRSL GRFKEPKLLE RTLGYLFDGT 840
841 VLNQDIYIPM QGMRAHQEGV EALWNWVKKN WDELVKRLPP GLSMLGSVVT LGTSGFTSMQ 900
901 KIDEIKKFFA TKSTKGFDQS LAQSLDTITS KAQWG |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
APE2 YDL241W |
| View Details | Ho Y, et al. (2002) |

|
ACS2 ADE3 ADE6 ADO1 ALA1 APE2 ARA1 BAT1 CDC10 CLU1 CYS3 DED81 DPS1 ERG10 ERG20 GCY1 GPH1 GRS1 HOM2 HYP2 ILS1 IMD3 KRS1 LSP1 MDH3 OLA1 PAB1 PFK2 PGM2 PMI40 PRP6 SAC6 SCP160 SOD1 SSZ1 THR4 TRR1 UBA1 YDL124W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
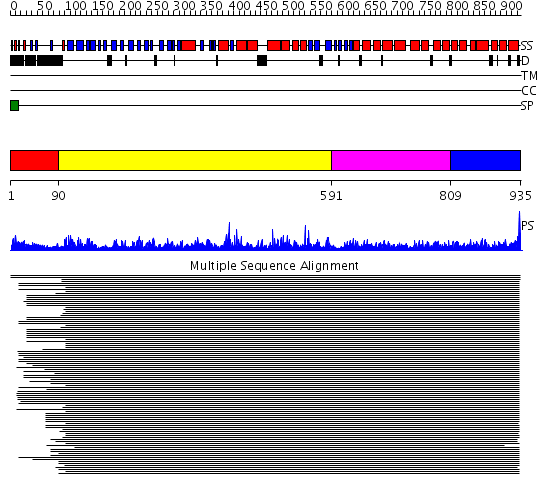
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | 1.303987 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [90..590] | 84.69897 | Leukotriene A4 hydrolase C-terminal domain; Leukotriene A4 hydrolase N-terminal domain; Leukotriene A4 hydrolase catalytic domain | |
| 3 | View Details | [591..808] | 1.228966 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [809..935] | 3.22199 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.74 |
Source: Reynolds et al. (2008)