













| General Information: |
|
| Name(s) found: |
HOC1 /
YJR075W
[SGD]
|
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 396 amino acids |
Gene Ontology: |
|
| Cellular Component: |
alpha-1,6-mannosyltransferase complex
[IDA |
| Biological Process: |
cell wall mannoprotein biosynthetic process
[TAS substituted mannan metabolic process [TAS |
| Molecular Function: |
transferase activity, transferring glycosyl groups
[ISS alpha-1,6-mannosyltransferase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKTTKRASS FRRLMIFAII ALISLAFGVR YLFHNSNATD LQKILQNLPK EISQSINSAN 60
61 NIQSSDSDLV QHFESLAQEI RHQQEVQAKQ FDKQRKILEK KIQDLKQTPP EATLRERIAM 120
121 TFPYDSHVKF PAFIWQTWSN DEGPERVQDI KGMWESKNPG FAHEVLNHDV INALVHHYFY 180
181 SIPEILETYE ALPSIILKID FFKYLILLVH GGVYADIDTF PVQPIPNWIP EELSPSDIGL 240
241 IVGVEEDAQR ADWRTKYIRR LQFGTWIIQA KPGHPVLREI ISRIIETTLQ RKRDDQLNVN 300
301 LRNDLNIMSW TGSGLWTDTI FTYFNDFMRS GVREKVTWKL FHNLNQPKLL SDVLVFPKFS 360
361 FNCPNQIDND DPHKKFYFIT HLASQFWKNT PKVEQK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
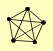
ANP1 HOC1 MNN10 MNN11 MNN9 |
| View Details | Gavin AC, et al. (2006) |
|
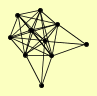
ANP1 HOC1 MNN10 MNN11 MNN9 |
| View Details | Qiu et al. (2008) |
|
ANP1 GNT1 HOC1 MNN10 MNN11 MNN5 MNN9 OCH1 VAN1 VRG4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
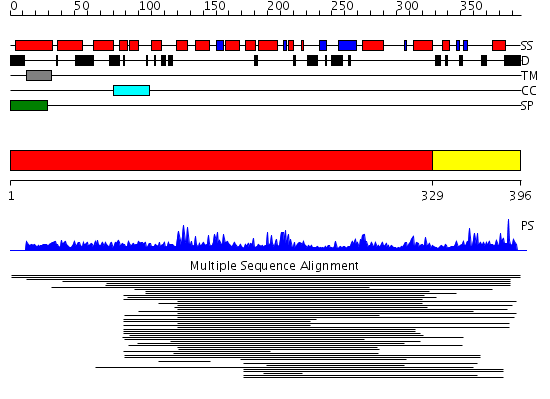
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..328] | 7.042971 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [329..396] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|