













| General Information: |
|
| Name(s) found: |
HSE1 /
YHL002W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 452 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endosome
[IDA |
| Biological Process: |
protein targeting to vacuole
[IMP |
| Molecular Function: |
protein binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSSAIKIRN ALLKATDPKL RSDNWQYILD VCDLVKEDPE DNGQEVMSLI EKRLEQQDAN 60
61 VILRTLSLTV SLAENCGSRL RQEISSKNFT SLLYALIESH SVHITLKKAV TDVVKQLSDS 120
121 FKDDPSLRAM GDLYDKIKRK APYLVQPNVP EKHNMSTQAD NSDDEELQKA LKMSLFEYEK 180
181 QKKLQEQEKE SAEVLPQQQQ QHQQQNQAPA HKIPAQTVVR RVRALYDLTT NEPDELSFRK 240
241 GDVITVLEQV YRDWWKGALR GNMGIFPLNY VTPIVEPSKE EIEKEKNKEA IVFSQKTTID 300
301 QLHNSLNAAS KTGNSNEVLQ DPHIGDMYGS VTPLRPQVTR MLGKYAKEKE DMLSLRQVLA 360
361 NAERSYNQLM DRAANAHISP PVPGPALYAG MTHANNTPVM PPQRQSYQSN EYSPYPSNLP 420
421 IQHPTNSANN TPQYGYDLGY SVVSQPPPGY EQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
HSE1 VPS27 |
| View Details | Krogan NJ, et al. (2006) |

|
HSE1 VPS27 |
| View Details | Gavin AC, et al. (2006) |

|
HSE1 VPS27 |
| View Details | Gavin AC, et al. (2002) |

|
HSE1 VPS27 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
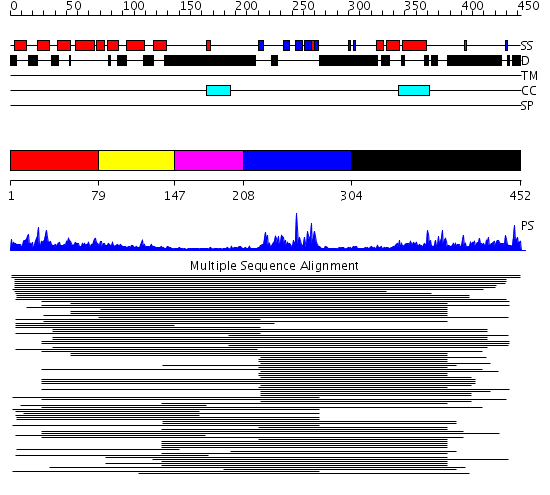
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..78] | 132.594777 | Tom1 protein | |
| 2 | View Details | [79..146] | 132.594777 | Tom1 protein | |
| 3 | View Details | [147..207] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [208..303] | 137.94696 | Growth factor receptor-bound protein 2 (GRB2), N- and C-terminal domains; Growth factor receptor-bound protein 2 (GRB2) | |
| 5 | View Details | [304..452] | 137.94696 | Growth factor receptor-bound protein 2 (GRB2), N- and C-terminal domains; Growth factor receptor-bound protein 2 (GRB2) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)