













| General Information: |
|
| Name(s) found: |
ICP55 /
YER078C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 511 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[NAS mitochondrion [IDA |
| Biological Process: |
proteolysis
[NAS |
| Molecular Function: |
X-Pro aminopeptidase activity
[ISS metalloendopeptidase activity [NAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLHRINPVRF SMQSCQRYFS KLVSPLEQHK SNTFTNRVRI PIEAGQPLHE TRPFLIKSGE 60
61 LTPGISALEY YERRIRLAET LPPKSCVILA GNDIQFASGA VFYPFQQEND LFYLSGWNEP 120
121 NSVMILEKPT DSLSDTIFHM LVPPKDAFAE KWEGFRSGVY GVQEIFNADE SASINDLSKY 180
181 LPKIINRNDF IYFDMLSTSN PSSSNFKHIK SLLDGSGNSN RSLNSIANKT IKPISKRIAE 240
241 FRKIKSPQEL RIMRRAGQIS GRSFNQAFAK RFRNERTLDS FLHYKFISGG CDKDAYIPVV 300
301 ATGSNSLCIH YTRNDDVMFD DEMVLVDAAG SLGGYCADIS RTWPNSGKFT DAQRDLYEAV 360
361 LNVQRDCIKL CKASNNYSLH DIHEKSITLM KQELKNLGID KVSGWNVEKL YPHYIGHNLG 420
421 LDVHDVPKVS RYEPLKVGQV ITIEPGLYIP NEESFPSYFR NVGIRIEDDI AIGEDTYTNL 480
481 TVEAVKEIDD LENVMQNGLS TKFEEDQVAP L |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
ADK1 ICP55 |
| View Details | Ho Y, et al. (2002) |
|
ARP2 CDC5 HHF1, HHF2 HHO1 ICP55 LOC1 MGM101 MUS81 NOP12 NUG1 RFA1 RPT2 RVB1 TAF14 TMA29 YRA1 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
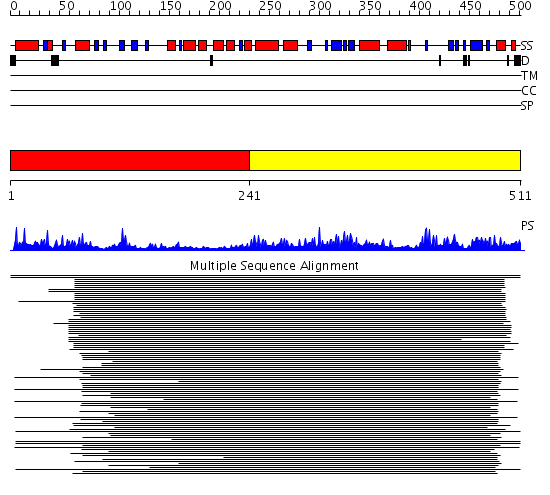
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..240] | 663.9794 | Aminopeptidase P; Aminopeptidase P, C-terminal domain | |
| 2 | View Details | [241..511] | 663.9794 | Aminopeptidase P; Aminopeptidase P, C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)