













| General Information: |
|
| Name(s) found: |
GIS1 /
YDR096W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 894 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mitochondrion [IDA |
| Biological Process: |
phospholipid metabolic process
[IMP ascospore wall assembly [IMP |
| Molecular Function: |
transcription factor activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEIKPVEVID GVPVFKPSMM EFANFQYFID EITKFGIENG IVKVIPPKEW LELLEGSPPA 60
61 ESLKTIQLDS PIQQQAKRWD KHENGVFSIE NEYDNKSYNL TQWKNLAESL DSRISQGDFN 120
121 DKTLKENCRV DSQQDCYDLA QLQILESDFW KTIAFSKPFY AVDENSSIFP YDLTLWNLNN 180
181 LPDSINSSNR RLLTGQSKCI FPWHLDEQNK CSINYLHFGA PKQWYSIPSA NTDQFLKILS 240
241 KEPSSNKENC PAFIRHQNII TSPDFLRKNN IKFNRVVQFQ HEFIITFPYC MYSGFNYGYN 300
301 FGESIEFILD QQAVVRKQPL KCGCGNKKEE RKSGPFSNLS YDSNESEQRG SITDNDNDLF 360
361 QKVRSFDELL NHSSQELQNL EDNKNPLFSN INMNRPQSSS LRSTTPNGVN QFLNMNQTTI 420
421 SRISSPLLSR MMDLSNIVEP TLDDPGSKFK RKVLTPQLPQ MNIPSNSSNF GTPSLTNTNS 480
481 LLSNITATST NPSTTTNGSQ NHNNVNANGI NTSAAASINN NISSTNNSAN NSSSNNNVST 540
541 VPSSMMHSST LNGTSGLGGD NDDNMLALSL ATLANSATAS PRLTLPPLSS PMNPNGHTSY 600
601 NGNMMNNNSG NGSNGSNSYS NGVTTAAATT TSAPHNLSIV SPNPTYSPNP LSLYLTNSKN 660
661 PLNSGLAPLS PSTSNIPFLK RNNVVTLNIS REASKSPISS FVNDYRSPLG VSNPLMYSST 720
721 INDYSNGTGI RQNSNNINPL DAGPSFSPLH KKPKILNGND NSNLDSNNFD YSFTGNKQES 780
781 NPSILNNNTN NNDNYRTSSM NNNGNNYQAH SSKFGENEVI MSDHGKIYIC RECNRQFSSG 840
841 HHLTRHKKSV HSGEKPHSCP RCGKRFKRRD HVLQHLNKKI PCTQEMENTK LAES |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
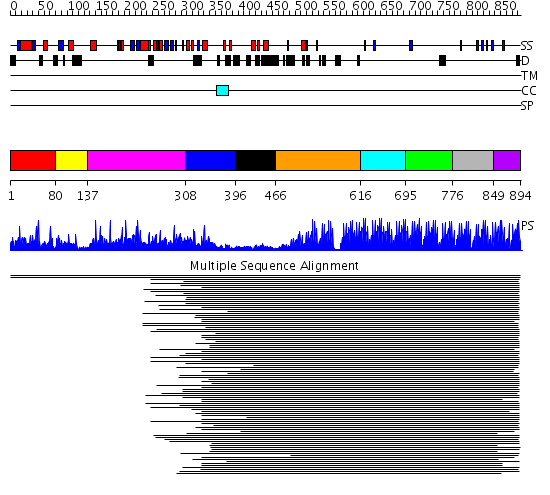
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..79] | 28.508638 | jmjN domain No confident structure predictions are available. | |
| 2 | View Details | [80..136] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [137..307] | 50.327902 | JmjC domain No confident structure predictions are available. | |
| 4 | View Details | [308..395] | 19.514997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [396..465] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [466..615] | 17.69897 | Five-finger GLI1 | |
| 7 | View Details | [616..694] | 89.24799 | ZIF268 | |
| 8 | View Details | [695..775] | 13.281994 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [776..848] | 2.69897 | Tramtrack protein (two zinc-finger peptide) | |
| 10 | View Details | [849..894] | 78.30103 | ADR1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)