













| General Information: |
|
| Name(s) found: |
egr-PB /
FBpp0087494
[FlyBase]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 409 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IDA][ISS]
|
| Biological Process: |
immune response
[IMP]
defense response to Gram-negative bacterium [IMP] induction of apoptosis [IGI][IMP] asymmetric protein localization involved in cell fate determination [IMP] apoptosis [NAS] apical protein localization [IMP] JNK cascade [IGI][IMP][NAS] |
| Molecular Function: |
protein binding
[IPI]
tumor necrosis factor receptor binding [IEA][ISS][NAS][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTAETLKPFI TPTSANDDGF PAKATSTATA QRRTRQLIPL VLGFIGLGLV VAILALTIWQ 60
61 TTRVSHLDKE LKSLKRVVDN LQQRLGINYL DEFDEFQKEY ENALIDYPKK VDGLTDEEDD 120
121 DDGDGLDSIA DDEDDDVSYS SVDDVGADYE DYTDMLNKLN NAHTGTTPTS ETTAEGEGET 180
181 DSASSASNDD NVFDDFTSYN AHKKKQERKS RSIADVRNEE QNIQGNHTEL QEKSSNEATS 240
241 KESPAPLHHR RRMHSRHRHL LVRKARSEDS RPAAHFHLSS RRRHQGSMGY HGDMYIGNDN 300
301 ERNSYQGHFQ TRDGVLTVTN TGLYYVYAQI CYNNSHDQNG FIVFQGDTPF LQCLNTVPTN 360
361 MPHKVHTCHT SGLIHLERNE RIHLKDIHND RNAVLREGNN RSYFGIFKV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
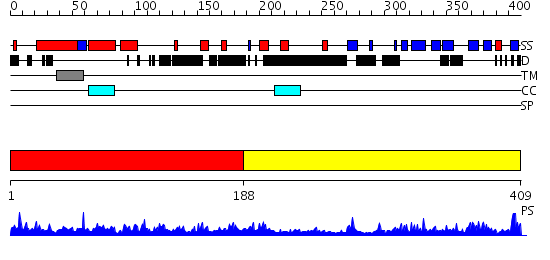
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..187] | 1.15 | Crystal structure of the periplasmic chaperone Skp | |
| 2 | View Details | [188..409] | 32.0 | Soluble part of TALL-1, sTALL-1 (BAFF) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|