













| General Information: |
|
| Name(s) found: |
Taf1-PB /
FBpp0089369
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2129 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[NAS]
transcription factor TFIID complex [IPI][ISS][NAS] |
| Biological Process: |
protein complex assembly
[NAS]
regulation of transcription, DNA-dependent [IEA][NAS] histone acetylation [IDA] regulation of transcription involved in G2/M-phase of mitotic cell cycle [IMP] regulation of cell cycle [NAS] positive regulation of transcription from RNA polymerase II promoter [IDA] transcription initiation from RNA polymerase II promoter [ISS] |
| Molecular Function: |
DNA binding
[NAS]
zinc ion binding [IEA] histone serine kinase activity [IDA] general RNA polymerase II transcription factor activity [ISS] protein kinase activity [NAS] sequence-specific DNA binding [IDA] transcription activator activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEMESDNSDD EGSIGNGLDL TGILFGNIDS EGRLLQDDDG EGRGGTGFDA ELRENIGSLS 60
61 KLGLDSMLLE VIDLKEAEPP SDDEEEEDAR PSAVSASEGM SAFDALKAGV KREDGAVKAQ 120
121 DDAIDYSDIT ELSEDCPRTP PEETSTYDDL EDAIPASKVE AKLTKDDKEL MPPPSAPMRS 180
181 GSGGGIEEPA KSNDASSPSD DSKSTDSKDA DRKLDTPLAD ILPSKYQNVD VRELFPDFRP 240
241 QKVLRFSRLF GPGKPTSLPQ IWRHVRKRRR KRNQSRDQKT TNTGGSDSPS DTEEPRKRGF 300
301 SLHYAAEPTP AECMSDDEDK LLGDFNSEDV RPEGPDNGEN SDQKPKVADW RFGPAQIWYD 360
361 MLEVPDSGEG FNYGFKTKAA STSSQPQLKD ERRVKSPEDD VEDPSIADDA FLMVSQLHWE 420
421 DDVVWDGNDI KAKVLQKLNS KTNAAGWLPS SGSRTAGAFS QPGKPSMPVG SGSSKQGSGA 480
481 SSKKAQQNAQ AKPAEAPDDT WYSLFPVENE ELIYYKWEDE VIWDAQQVSK VPKPKVLTLD 540
541 PNDENIILGI PDDIDPSKIN KSTGPPPKIK IPHPHVKKSK ILLGKAGVIN VLAEDTPPPP 600
601 PKSPDRDPFN ISNDTYYTPK TEPTLRLKVG GNLIQHSTPV VELRAPFVPT HMGPMKLRAF 660
661 HRPPLKKYSH GPMAQSIPHP VFPLLKTIAK KAKQREVERI ASGGGDVFFM RNPEDLSGRD 720
721 GDIVLAEFCE EHPPLINQVG MCSKIKNYYK RKAEKDSGPQ DFVYGEVAFA HTSPFLGILH 780
781 PGQCIQAIEN NMYRAPIYPH KMAHNDFLVI RTRNNYWIRS VNSIYTVGQE CPLYEVPGPN 840
841 SKRANNFTRD FLQVFIYRLF WKSRDNPRRI RMDDIKQAFP AHSESSIRKR LKQCADFKRT 900
901 GMDSNWWVIK PEFRLPSEEE IRAMVSPEQC CAYFSMIAAE QRLKDAGYGE KFLFAPQEDD 960
961 DEEAQLKLDD EVKVAPWNTT RAYIQAMRGK CLLQLSGPAD PTGCGEGFSY VRVPNKPTQT1020
1021 KEEQESQPKR SVTGTDADLR RLPLQRAKEL LRQFKVPEEE IKKLSRWEVI DVVRTLSTEK1080
1081 AKAGEEGMDK FSRGNRFSIA EHQERYKEEC QRIFDLQNRV LASSEVLSTD EAESSASEES1140
1141 DLEELGKNLE NMLSNKKTST QLSREREELE RQELLRQLDE EHGGPSGSGG AKGAKGKDDP1200
1201 GQQMLATNNQ GRILRITRTF RGNDGKEYTR VETVRRQPVI DAYIKIRTTK DEQFIKQFAT1260
1261 LDEQQKEEMK REKRRIQEQL RRIKRNQERE RLAQLAQNQK LQPGGMPTSL GDPKSSGGHS1320
1321 HKERDSGYKE VSPSRKKFKL KPDLKLKCGA CGQVGHMRTN KACPLYSGMQ SSLSQSNPSL1380
1381 ADDFDEQSEK EMTMDDDDLV NVDGTKVTLS SKILKRHGGD DGKRRSGSSS GFTLKVPRDA1440
1441 MGKKKRRVGG DLHCDYLQRH NKTANRRRTD PVVVLSSILE IIHNELRSMP DVSPFLFPVS1500
1501 AKKVPDYYRV VTKPMDLQTM REYIRQRRYT SREMFLEDLK QIVDNSLIYN GPQSAYTLAA1560
1561 QRMFSSCFEL LAEREDKLMR LEKAINPLLD DDDQVALSFI FDKLHSQIKQ LPESWPFLKP1620
1621 VNKKQVKDYY TVIKRPMDLE TIGKNIEAHR YHSRAEYLAD IELIATNCEQ YNGSDTRYTK1680
1681 FSKKILEYAQ TQLIEFSEHC GQLENNIAKT QERARENAPE FDEAWGNDDY NFDRGSRASS1740
1741 PGDDYIDVEG HGGHASSSNS IHRSMGAEAG SSHTAPAVRK PAPPGPGEVK RGRGRPRKQR1800
1801 DPVEEVKSQN PVKRGRGRPR KDSLASNMSH TQAYFLDEDL QCSTDDEDDD EEEDFQEVSE1860
1861 DENNAASILD QGERINAPAD AMDGMFDPKN IKTEIDLEAH QMAEEPIGED DSQQVAEAMV1920
1921 QLSGVGGYYA QQQQDESMDV DPNYDPSDFL AMHKQRQSLG EPSSLQGAFT NFLSHEQDDN1980
1981 GPYNPAEAST SAASGADLGM DASMAMQMAP EMPVNTMNNG MGIDDDLDIS ESDEEDDGSR2040
2041 VRIKKEVFDD GDYALQHQQM GQAASQSQIY MVDSSNEPTT LDYQQPPQLD FQQVQEMEQL2100
2101 QHQVMPPMQS EQLQQQQTPQ GDNDYAWTF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
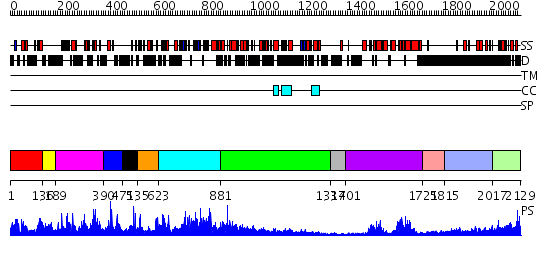
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..135] | 22.09691 | TAF(II)230 TBP-binding fragment | |
| 2 | View Details | [136..188] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [189..389] | 1.020949 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [390..470] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [471..534] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [535..622] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [623..880] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [881..1336] | 4.522879 | alpha-actinin | |
| 9 | View Details | [1337..1400] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1401..1724] | 49.045757 | TAFII250 double bromodomain module | |
| 11 | View Details | [1725..1814] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1815..2016] | 1.242988 | View MSA. No confident structure predictions are available. | |
| 13 | View Details | [2017..2129] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 8 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)