













| General Information: |
|
| Name(s) found: |
dnc-PG /
FBpp0070496
[FlyBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 814 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATEAEGEEF DVDPMDEDDE DQTYDRETEE FYSNIQDAAG TGSSSRSKRS SLFSRSDSSA 60
61 TTTSSSGGGT FTGGKRRSAA SILSSSMCSD LMTSDRRSST ATEYSVKSVT TGNTSQRRSS 120
121 GRIRRYVSRM TIAGARRRTT GSFDVENGQG ARSPLEGGSP SAGLVLQNLP QRRESFLYRS 180
181 DSDFEMSPKS MSRNSSIASE SHGEDLIVTP FAQILASLRS VRNNLLSLTN VPASNKSRRP 240
241 NQSSSASRSG NPPGAPLSQG EEAYTRLATD TIEELDWCLD QLETIQTHRS VSDMASLKFK 300
301 RMLNKELSHF SESSRSGNQI SEYICSTFLD KQQEFDLPSL RVEDNPELVA ANAAAGQQSA 360
361 GQYARSRSPR GPPMSQISGV KRPLSHTNSF TGERLPTFGV ETPRENELGT LLGELDTWGI 420
421 QIFSIGEFSV NRPLTCVAYT IFQSRELLTS LMIPPKTFLN FMSTLEDHYV KDNPFHNSLH 480
481 AADVTQSTNV LLNTPALEGV FTPLEVGGAL FAACIHDVDH PGLTNQFLVN SSSELALMYN 540
541 DESVLENHHL AVAFKLLQNQ GCDIFCNMQK KQRQTLRKMV IDIVLSTDMS KHMSLLADLK 600
601 TMVETKKVAG SGVLLLDNYT DRIQVLENLV HCADLSNPTK PLPLYKRWVA LLMEEFFLQG 660
661 DKERESGMDI SPMCDRHNAT IEKSQVGFID YIVHPLWETW ADLVHPDAQD ILDTLEENRD 720
721 YYQSMIPPSP PPSGVDENPQ EDRIRFQVTL EESDQENLAE LEEGDESGGE STTTGTTGTT 780
781 AASALSGAGG GGGGGGGMAP RTGGCQNQPQ HGGM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
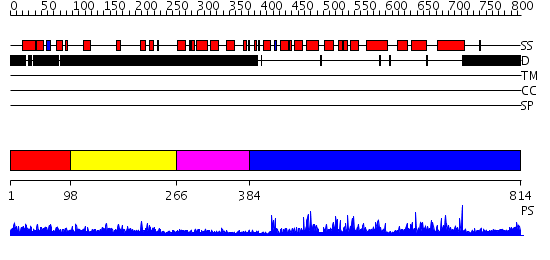
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..97] | 1.082983 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [98..265] | 6.145997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [266..383] | 1.200989 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [384..814] | 136.0 | No description for 2fm0A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)