













| General Information: |
|
| Name(s) found: |
SMG7
[HGNC (HUGO)]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 1137 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLQSAQYLR QAEVLKADMT DSKLGPAEVW TSRQALQDLY QKMLVTDLEY ALDKKVEQDL 60
61 WNHAFKNQIT TLQGQAKNRA NPNRSEVQAN LSLFLEAASG FYTQLLQELC TVFNVDLPCR 120
121 VKSSQLGIIS NKQTHTSAIV KPQSSSCSYI CQHCLVHLGD IARYRNQTSQ AESYYRHAAQ 180
181 LVPSNGQPYN QLAILASSKG DHLTTIFYYC RSIAVKFPFP AASTNLQKAL SKALESRDEV 240
241 KTKWGVSDFI KAFIKFHGHV YLSKSLEKLS PLREKLEEQF KRLLFQKAFN SQQLVHVTVI 300
301 NLFQLHHLRD FSNETEQHTY SQDEQLCWTQ LLALFMSFLG ILCKCPLQNE SQEESYNAYP 360
361 LPAVKVSMDW LRLRPRVFQE AVVDERQYIW PWLISLLNSF HPHEEDLSSI SATPLPEEFE 420
421 LQGFLALRPS FRNLDFSKGH QGITGDKEGQ QRRIRQQRLI SIGKWIADNQ PRLIQCENEV 480
481 GKLLFITEIP ELILEDPSEA KENLILQETS VIESLAADGS PGLKSVLSTS RNLSNNCDTG 540
541 EKPVVTFKEN IKTREVNRDQ GRSFPPKEVR RDYSKGITVT KNDGKKDNNK RKTETKKCTL 600
601 EKLQETGKQN VAVQVKSQTE LRKTPVSEAR KTPVTQTPTQ ASNSQFIPIH HPGAFPPLPS 660
661 RPGFPPPTYV IPPPVAFSMG SGYTFPAGVS VPGTFLQPTA HSPAGNQVQA GKQSHIPYSQ 720
721 QRPSGPGPMN QGPQQSQPPS QQPLTSLPAQ PTAQSTSQLQ VQALTQQQQS PTKAVPALGK 780
781 SPPHHSGFQQ YQQADASKQL WNPPQVQGPL GKIMPVKQPY YLQTQDPIKL FEPSLQPPVM 840
841 QQQPLEKKMK PFPMEPYNHN PSEVKVPEFY WDSSYSMADN RSVMAQQANI DRRGKRSPGV 900
901 FRPEQDPVPR MPFEKSLLEK PSELMSHSSS FLSLTGFSLN QERYPNNSMF NEVYGKNLTS 960
961 SSKAELSPSM APQETSLYSL FEGTPWSPSL PASSDHSTPA SQSPHSSNPS SLPSSPPTHN1020
1021 HNSVPFSNFG PIGTPDNRDR RTADRWKTDK PAMGGFGIDY LSATSSSESS WHQASTPSGT1080
1081 WTGHGPSMED SSAVLMESLK SIWSSSMMHP GPSALEQLLM QQKQKQQRGQ GTMNPPH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
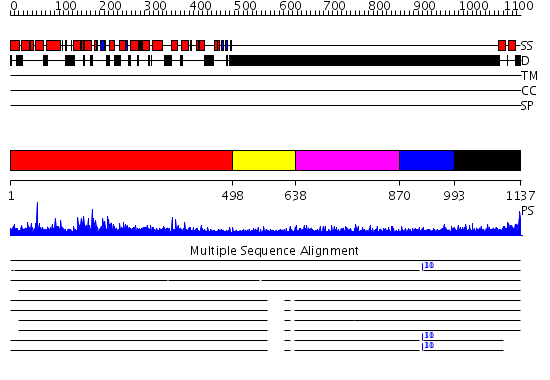
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..497] | 1000.0 | Crystal structure of the N-terminal domain of human SMG7 | |
| 2 | View Details | [498..637] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [638..869] | 2.281997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [870..992] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [993..1137] | 1.05 | Chondroitinase B |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)