













| General Information: |
|
| Name(s) found: |
E2F8
[HGNC (HUGO)]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 867 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MENEKENLFC EPHKRGLMKT PLKESTTANI VLAEIQPDFG PLTTPTKPKE GSQGEPWTPT 60
61 ANLKMLISAV SPEIRNRDQK RGLFDNRSGL PEAKDCIHEH LSGDEFEKSQ PSRKEKSLGL 120
121 LCHKFLARYP NYPNPAVNND ICLDEVAEEL NVERRRIYDI VNVLESLHMV SRLAKNRYTW 180
181 HGRHNLNKTL GTLKSIGEEN KYAEQIMMIK KKEYEQEFDF IKSYSIEDHI IKSNTGPNGH 240
241 PDMCFVELPG VEFRAASVNS RKDKSLRVMS QKFVMLFLVS TPQIVSLEVA AKILIGEDHV 300
301 EDLDKSKFKT KIRRLYDIAN VLSSLDLIKK VHVTEERGRK PAFKWTGPEI SPNTSGSSPV 360
361 IHFTPSDLEV RRSSKENCAK NLFSTRGKPN FTRHPSLIKL VKSIESDRRK INSAPSSPIK 420
421 TNKAESSQNS APFPSKMAQL AAICKMQLEE QSSESRQKVK VQLARSGPCK PVAPLDPPVN 480
481 AEMELTAPSL IQPLGMVPLI PSPLSSAVPL ILPQAPSGPS YAIYLQPTQA HQSVTPPQGL 540
541 SPTVCTTHSS KATGSKDSTD ATTEKAANDT SKASASTRPG SLLPAPERQG AKSRTREPAG 600
601 ERGSKRASML EDSGSKKKFK EDLKGLENVS ATLFPSGYLI PLTQCSSLGA ESILSGKENS 660
661 SALSPNHRIY SSPIAGVIPV TSSELTAVNF PSFHVTPLKL MVSPTSVAAV PVGNSPALAS 720
721 SHPVPIQNPS SAIVNFTLQH LGLISPNVQL SASPGSGIVP VSPRIESVNV APENAGTQQG 780
781 RATNYDSPVP GQSQPNGQSV AVTGAQQPVP VTPKGSQLVA ESFFRTPGGP TKPTSSSCMD 840
841 FEGANKTSLG TLFVPQRKLE VSTEDVH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
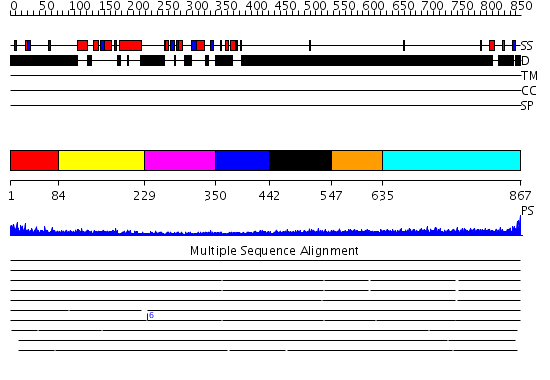
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..83] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [84..228] | 13.69897 | Cell cycle transcription factor E2F-4 | |
| 3 | View Details | [229..349] | 2.38 | Cell cycle transcription factor E2F-4 | |
| 4 | View Details | [350..441] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [442..546] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [547..634] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [635..867] | 4.245994 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)