













| General Information: |
|
| Name(s) found: |
gi|36950991
[NCBI NR]
gi|119584618 [NCBI NR] gi|119584617 [NCBI NR] |
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 615 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSPSPRIQI ISTDSAVASP QRIQGSEPAS GPLSVFTSLN KEKIVTDQQT GQKIQIVTAV 60
61 DASGSPKQQF ILTSPDGAGT GKVILASPET SSAKQLIFTT SDNLVPGRIQ IVTDSASVER 120
121 LLGKTDVQRP QVVEYCVVCG DKASGRHYGA VSCEGCKGFF KRSVRKNLTY SCRSNQDCII 180
181 NKHHRNRCQF CRLKKCLEMG MKMESVQSER KPFDVQREKP SNCAASTEKI YIRKDLRSPL 240
241 IATPTFVADK DGARQTGLLD PGMLVNIQQP LIREDGTVLL ATDSKAETSQ GALGTLANVV 300
301 TSLANLSESL NNGDTSEIQP EDQSASEITR AFDTLAKALN TTDSSSSPSL ADGIDTSGGG 360
361 SIHVISRDQS TPIIEVEGPL LSDTHVTFKL TMPSPMPEYL NVHYICESAS RLLFLSMHWA 420
421 RSIPAFQALG QDCNTSLVRA CWNELFTLGL AQCAQVMSLS TILAAIVNHL QNSIQEDKLS 480
481 GDRIKQVMEH IWKLQEFCNS MAKLDIDGYE YAYLKAIVLF SPDHPGLTST SQIEKFQEKA 540
541 QMELQDYVQK TYSEDTYRLA RILVRLPALR LMSSNITEEL FFTGLIGNVS IDSIIPYILK 600
601 METAEYNGQI TGASL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
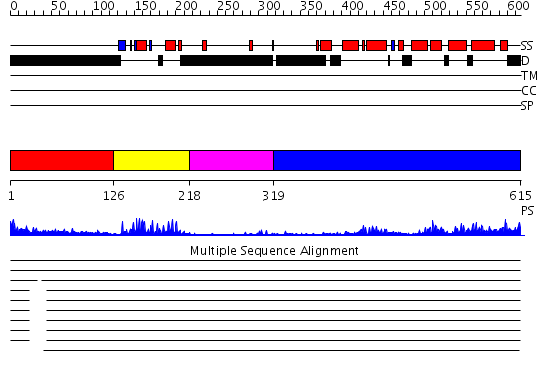
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..125] | 13.291997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [126..217] | 24.69897 | Human Liver Receptor Homologue DNA-Binding Domain (hLRH-1 DBD) in Complex with dsDNA from the hCYP7A1 Promoter | |
| 3 | View Details | [218..318] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [319..615] | 51.69897 | LIGAND-BINDING DOMAIN OF THE HUMAN NUCLEAR RECEPTOR RXR-ALPHA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)