













| General Information: |
|
| Name(s) found: |
TUBA3D
[HGNC (HUGO)]
TUBA3C [HGNC (HUGO)] |
| Description(s) found:
Found 45 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 450 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK 60
61 HVPRAVFVDL EPTVVDEVRT GTYRQLFHPE QLITGKEDAA NNYARGHYTI GKEIVDLVLD 120
121 RIRKLADLCT GLQGFLIFHS FGGGTGSGFA SLLMERLSVD YGKKSKLEFA IYPAPQVSTA 180
181 VVEPYNSILT THTTLEHSDC AFMVDNEAIY DICRRNLDIE RPTYTNLNRL IGQIVSSITA 240
241 SLRFDGALNV DLTEFQTNLV PYPRIHFPLA TYAPVISAEK AYHEQLSVAE ITNACFEPAN 300
301 QMVKCDPRHG KYMACCMLYR GDVVPKDVNA AIATIKTKRT IQFVDWCPTG FKVGINYQPP 360
361 TVVPGGDLAK VQRAVCMLSN TTAIAEAWAR LDHKFDLMYA KRAFVHWYVG EGMEEGEFSE 420
421 AREDLAALEK DYEEVGVDSV EAEAEEGEEY |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample pIC145 from april 2005 | Okada, M., et al. (2006) | |
| View Run | Sample pIC146 from april 2005 | Okada, M., et al. (2006) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
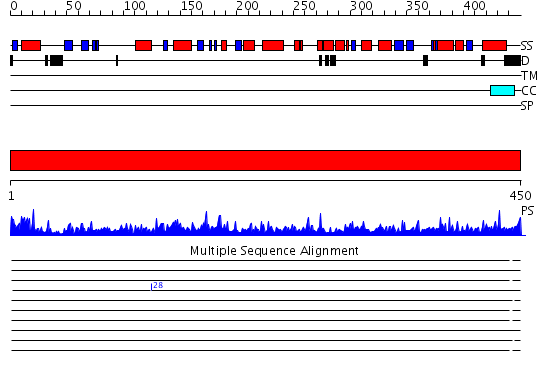
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..450] | 1000.0 | Tubulin-colchicine-vinblastine: stathmin-like domain complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.69 |
Source: Reynolds et al. (2008)