













| General Information: |
|
| Name(s) found: |
gi|151942718
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae YJM789 |
| Length: | 244 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSMSMGVRGL ALRSVSKTLF SQGVRCPSMV IGARYMSSTP EKQTDPKAKA NSIINAIPGN 60
61 NILTKTGVLG TSAAAVIYAI SNELYVINDE SILLLTFLGF TGLVAKYLAP AYKDFADARM 120
121 KKVSDVLNAS RNKHVEAVKD RIDSVSQLQN VAETTKVLFD VSKETVELES EAFELKQKVE 180
181 LAHEAKAVLD SWVRYEASLR QLEQRQLAKS VISRVQSELG NPKFQEKVLQ QSISEIEQLL 240
241 SKLK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
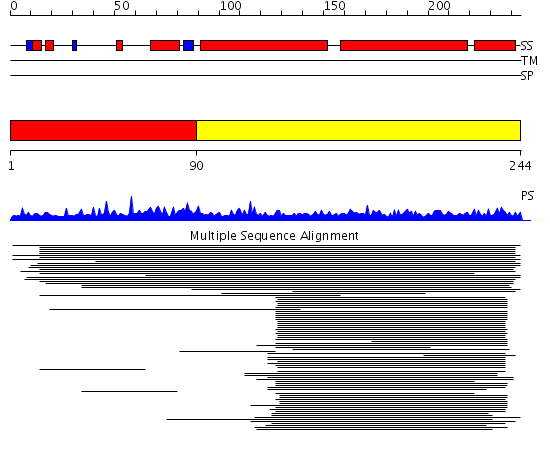
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | 2.020997 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [90..244] | 28.049997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions |
| 1 | |
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|