













| General Information: |
|
| Name(s) found: |
gi|151941546
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae YJM789 |
| Length: | 704 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDEHLISTIN KLQDALAPLG GGSQSPIDLP QITVVGSQSS GKSSVLENIV GRDFLPRGTG 60
61 IVTRRPLVLQ LINRRPKKSE HAKVNQTANE LIDLNINDDD KKKDESGKHQ NEGQSEDNKE 120
121 EWGEFLHLPG KKFYNFDEIR KEIVKETDKV TGANSGISSV PINLRIYSPH VLTLTLVDLP 180
181 GLTKVPVGDQ PPDIERQIKD MLLKYISKPN AIILSVNAAN TDLANSDGLK LAREVDPEGT 240
241 RTIGVLTKVD LMDQGTDVID ILAGRVIPLR YGYIPVINRG QKDIEHKKTI REALENERKF 300
301 FENHPSYSSK AHYCGTPYLA KKLNSILLHH IRQTLPEIKA KIEATLKKYQ NELINLGPET 360
361 MDSASSVVLS MITDFSNEYA GILDGEAKEL SSQELSGGAR ISYVFHETFK NGVDSLDPFD 420
421 QIKDSDIRTI MYNSSGSAPS LFVGTEAFEV LVKQQIRRFE EPSLRLVTLV FDELVRMLKQ 480
481 IISQPKYSRY PALREAISNQ FIQFLKDATI PTNEFVVDII KAEQTYINTA HPDLLKGSQA 540
541 MVMVEEKLHP RQVAVDPKTG KPLPTQPSSS KAPVMEEKSG FFGGFFSTKN KKKLAALESP 600
601 PPVLKATGQM TERETMETEV IKLLISSYFS IVKRTIADII PKALMLKLIV KSKTDIQKVL 660
661 LEKLYGKQDI EELTKENDIT IQRRKECKKM VEILRNASQI VSSV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
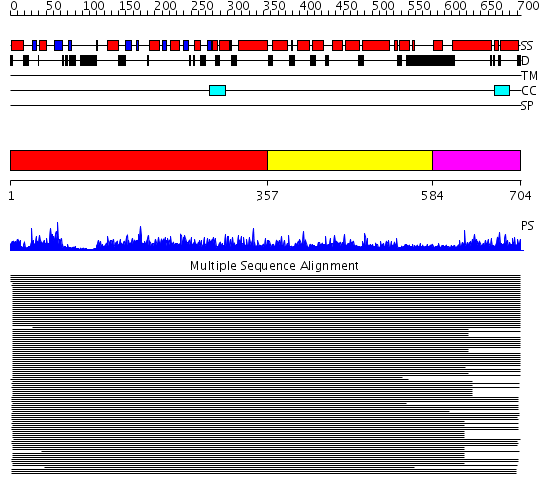
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..356] | 1185.0 | Dynamin G domain | |
| 2 | View Details | [357..583] | 42.69897 | Dynamin central region No confident structure predictions are available. | |
| 3 | View Details | [584..704] | 40.173925 | Dynamin GTPase effector domain No confident structure predictions are available. | |
| 4 | View Details | [600..704] | 40.187087 | No description for PF02212.9 was found. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)