













| General Information: |
|
| Name(s) found: |
dhs-30 /
CE37275
[WormBase]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 311 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
estrogen biosynthetic process
[IEA tetrahydrobiopterin biosynthetic process [IEA poly-hydroxybutyrate biosynthetic process [IEA metabolic process [IEA fatty acid biosynthetic process [IEA |
| Molecular Function: |
estradiol 17-beta-dehydrogenase activity
[IEA sepiapterin reductase activity [IEA 3-oxoacyl-[acyl-carrier-protein] reductase activity [IEA oxidoreductase activity [IEA NAD or NADH binding [IEA 3-hydroxybutyrate dehydrogenase activity [IEA acetoacetyl-CoA reductase activity [IEA 2-deoxy-D-gluconate 3-dehydrogenase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVPVKLQEML KEWAPALVVP LSLYVAYKLL NRIIPGAHNL PKLDVKNKVV VITGASSGLG 60
61 KSLAFELYKR GAQVILLARS TEKLKEICEE LKETFPLNQN EPIYYYFDIT DSEQAPWAEI 120
121 PRVDILINNA GMSNRGSCQD TTMEIHRQAM ETNYFGHVHV TQALLSKLSP DGCIVVTSSI 180
181 QGKVAIPYRG SYGASKHALQ GYFDCLRAEH KNLHILVVSA GYINTGFGSR ALDPSGKVVG 240
241 VEDENQKKGY TPEHCARMIS DAIRDRKTDF DMAPFDARIA VFLRYFWPTL LNYALYIRGT 300
301 KDQWAPKNKK E |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
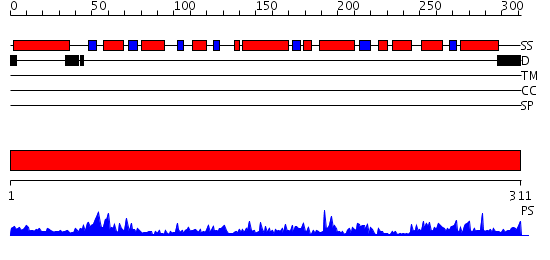
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..311] | 55.69897 | Crystal structure of peroxisomal trans 2-enoyl CoA reductase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)