













| General Information: |
|
| Name(s) found: |
CE36953
[WormBase]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 626 amino acids |
Gene Ontology: |
|
| Cellular Component: |
integral to membrane
[IEA |
| Biological Process: |
carbohydrate metabolic process
[IEA chitin catabolic process [IEA |
| Molecular Function: |
chitinase activity
[IEA chitin binding [IEA hydrolase activity, hydrolyzing O-glycosyl compounds [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRFVMWMWLF VVWITVLFGV SNGGILDRSC GRRRVGYITS WGKHPFRDDQ AEKLTHLVFA 60
61 FFVVDSDGSV KLEGDAAKAR LEHVKEVASR HPDLKLLYAV GGWENSQYFS VLTADHSRRS 120
121 ILISNFVKVI KEYGFDGVDI DWEYPVTGGA VEGTPADRRN YVNLMRELRN ELRDLESETG 180
181 KSYLISFAGA AGHWVLKPGY DLQQLMKYCD FVNVMSYDYF GAWASKWGAY TGPPAPLQFA 240
241 MPKKFSGRMN VHATMKDYSC QIKATDKINM GVPFYGRFWK NVGDAVDSTD DMWRTATATN 300
301 SEGTKFEGGD VQWRDLHEKF DTTKTKFHSG SKTPFIWLSE QKTFVGYENA ESLKHKVDYI 360
361 VENNIGGVMI WAIDFDDDQG TLLNSAAAES ICTTSTKSFN YKCSPVDDKR WWTYDDNEEL 420
421 AGMCGKSSPL IDGYYPVCDP DDPGHACCGK YGYCGSGAEF CSCPECIDYG ADPNLILKEP 480
481 VKPSQKITWY TSDAGEGKRG RCGRDVPPLE GEAPTCNPDD ANAHCCSNGG YCGNSKEHCE 540
541 CNGCIDFAKQ RDFKYKPLEW WTFSENPANV GRCGYNAPRL STGKIPKCDP DSESYCCSNS 600
601 GYCGKGEQYC SCLGCADFKA NPAFEY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
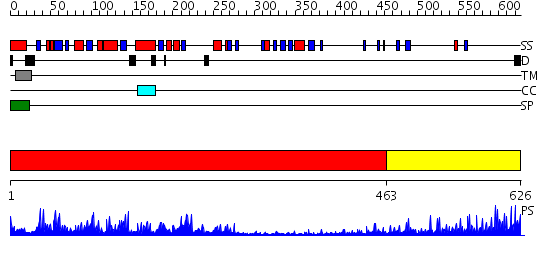
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..462] | 83.39794 | Specificity and affnity of natural product cyclopentapeptide inhibitor Argadin against human chitinase | |
| 2 | View Details | [463..626] | 9.30103 | Low density lipoprotein (LDL) receptor YWTD domain; Low density lipoprotein (LDL) receptor, different EGF domains; Ligand-binding domain of low-density lipoprotein receptor |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|