













| General Information: |
|
| Name(s) found: |
gi|30794015
[NCBI NR]
gi|15237923 [NCBI NR] gi|110742646 [NCBI NR] gi|10177085 [NCBI NR] |
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 746 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA][ISS]
|
| Biological Process: |
proteolysis
[IEA][ISS]
|
| Molecular Function: |
serine-type peptidase activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADKDVPFGV EDIVQTPLPG YVAPTAVSFS PDDSLITYLF SPEKNLKRRV YAFDVNKGES 60
61 NLVFSPPDGG VDESNISPEE KLRRERLRER GLGVTRYEWV KTNSKMRFIV VPLPAGVYMK 120
121 DLSSSPNPEL IVPSSPTSPI IDPRLSPNGL FLAYVRESEL HVLNLLKNQT QQLTSGANGS 180
181 TLSHGLAEYI AQEEMDRRNG YWWSLDSKFI AYTEVDSSQI PLFRIMHQGK RSVGSEAQED 240
241 HAYPFAGALN STLRLGVVSS AGGGKTTWMN LVCGGRGNTE DEYLGRVNWL PGNVLIVQVL 300
301 NRSQSKLKII SFDINTGQGN VLLTEESDTW VTLHDCFTPL EKVPSSRGSG GFIWASERTG 360
361 FRHLYLYESD GTCLGAITSG EWMVEQIAGV NEPMSLVYFT GTLDGPLETN LYCAKLEAGN 420
421 TSRCQPMRLT HGKGKHIVVL DHQMKNFVDI HDSVDSPPRV SLCSLSDGTV LKILYEQTSP 480
481 IQILKSLKLE PPEFVQIQAN DGKTTLYGAV YKPDSSKFGP PPYKTMINVY GGPSVQLVYD 540
541 SWINTVDMRT QYLRSRGILV WKLDNRGTAR RGLKFESWMK HNCGYVDAED QVTGAKWLIE 600
601 QGLAKPDHIG VYGWSYGGYL SATLLTRYPE IFNCAVSGAP VTSWDGYDSF YTEKYMGLPT 660
661 EEERYLKSSV MHHVGNLTDK QKLMLVHGMI DENVHFRHTA RLVNALVEAG KRYELLIFPD 720
721 ERHMPRKKKD RIYMEQRIWE FIEKNL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
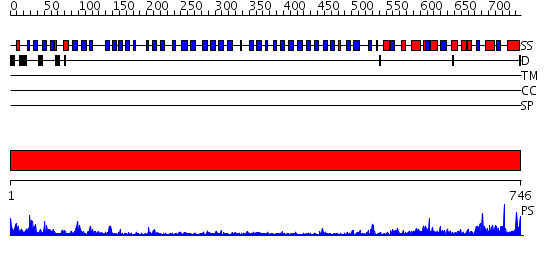
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..746] | 79.39794 | Crystal structure of HIV-1 Tat derived nonapeptides Tat(1-9) bound to the active site of Dipeptidyl peptidase IV (CD26) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)