













| General Information: |
|
| Name(s) found: |
ANXD1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 317 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] cell wall [IDA] mitochondrion [IDA] apoplast [IDA] membrane [IDA] cytosol [IDA] vacuole [IDA] plasma membrane [IDA] thylakoid [IDA] |
| Biological Process: |
response to cold
[IEP]
response to oxidative stress [IGI] response to heat [IEP] response to abscisic acid stimulus [IEP][IMP] response to salt stress [IEP] response to osmotic stress [IEP][IMP] response to cadmium ion [IEP] response to water deprivation [IEP] |
| Molecular Function: |
peroxidase activity
[IDA]
ATP binding [IDA] zinc ion binding [IDA] protein homodimerization activity [IDA] copper ion binding [IDA] calcium ion binding [ISS] calcium-dependent phospholipid binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATLKVSDSV PAPSDDAEQL RTAFEGWGTN EDLIISILAH RSAEQRKVIR QAYHETYGED 60
61 LLKTLDKELS NDFERAILLW TLEPGERDAL LANEATKRWT SSNQVLMEVA CTRTSTQLLH 120
121 ARQAYHARYK KSLEEDVAHH TTGDFRKLLV SLVTSYRYEG DEVNMTLAKQ EAKLVHEKIK 180
181 DKHYNDEDVI RILSTRSKAQ INATFNRYQD DHGEEILKSL EEGDDDDKFL ALLRSTIQCL 240
241 TRPELYFVDV LRSAINKTGT DEGALTRIVT TRAEIDLKVI GEEYQRRNSI PLEKAITKDT 300
301 RGDYEKMLVA LLGEDDA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
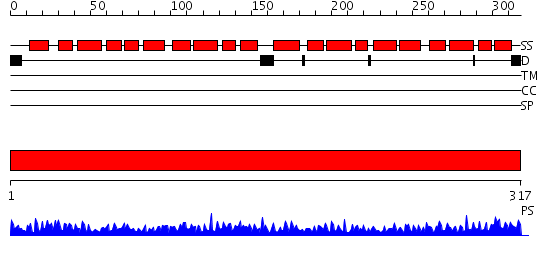
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..317] | 101.0 | X-RAY STRUCTURE OF ANNEXIN FROM ARABIDOPSIS THALIANA GENE AT1G35720 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)