













| General Information: |
|
| Name(s) found: |
GSTF9_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 215 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] cytoplasm [NAS] apoplast [IDA] vacuole [IDA] plasma membrane [IDA] thylakoid [IDA] |
| Biological Process: |
toxin catabolic process
[TAS]
response to zinc ion [IEP] response to cadmium ion [IDA] defense response [IEP] defense response to bacterium [IEP] |
| Molecular Function: |
glutathione peroxidase activity
[IDA]
copper ion binding [IDA] glutathione binding [IDA] glutathione transferase activity [IDA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLKVYGPHF ASPKRALVTL IEKGVAFETI PVDLMKGEHK QPAYLALQPF GTVPAVVDGD 60
61 YKIFESRAVM RYVAEKYRSQ GPDLLGKTVE DRGQVEQWLD VEATTYHPPL LNLTLHIMFA 120
121 SVMGFPSDEK LIKESEEKLA GVLDVYEAHL SKSKYLAGDF VSLADLAHLP FTDYLVGPIG 180
181 KAYMIKDRKH VSAWWDDISS RPAWKETVAK YSFPA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
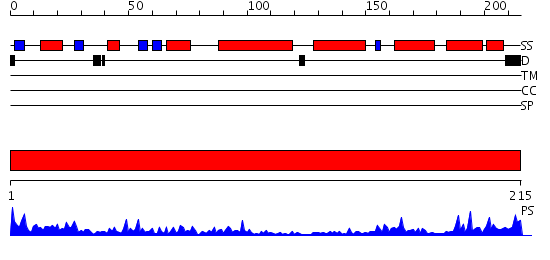
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..215] | 46.154902 | STRUCTURE OF GLUTATHIONE S-TRANSFERASE III IN APO FORM |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)