













| General Information: |
|
| Name(s) found: |
CAMK2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 599 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
protein amino acid phosphorylation
[IEA]
|
| Molecular Function: |
ATP binding
[IEA]
calcium-dependent protein serine/threonine phosphatase activity [ISS] calcium ion binding [ISS] protein kinase activity [IEA] protein serine/threonine kinase activity [IEA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGGCTSKPSS SVKPNPYAPK DAVLQNDDST PAHPGKSPVR SSPAVKASPF FPFYTPSPAR 60
61 HRRNKSRDGG GGESKSVTST PLRQLARAFH PPSPARHIRD VLRRRKEKKE AALPAARQQK 120
121 EEEEREEVGL DKRFGFSKEL QSRIELGEEI GRGHFGYTCS AKFKKGELKD QEVAVKVIPK 180
181 SKMTSAISIE DVRREVKILR ALSGHQNLVQ FYDAFEDNAN VYIVMELCGG GELLDRILAR 240
241 GGKYSEDDAK AVLIQILNVV AFCHLQGVVH RDLKPENFLY TSKEENSMLK VIDFGLSDFV 300
301 RPDERLNDIV GSAYYVAPEV LHRSYTTEAD VWSIGVIAYI LLCGSRPFWA RTESGIFRAV 360
361 LKADPSFDEP PWPSLSFEAK DFVKRLLYKD PRKRMTASQA LMHPWIAGYK KIDIPFDILI 420
421 FKQIKAYLRS SSLRKAALMA LSKTLTTDEL LYLKAQFAHL APNKNGLITL DSIRLALATN 480
481 ATEAMKESRI PDFLALLNGL QYKGMDFEEF CAASISVHQH ESLDCWEQSI RHAYELFEMN 540
541 GNRVIVIEEL ASELGVGSSI PVHTILNDWI RHTDGKLSFL GFVKLLHGVS TRQSLAKTR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
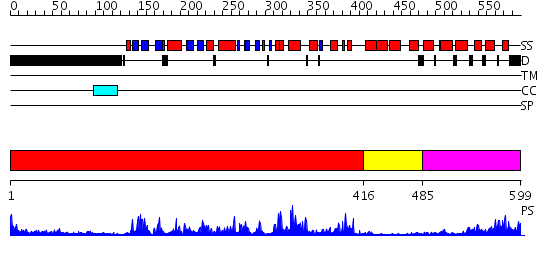
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..415] | 61.0 | Abl tyrosine kinase, SH3 domain; Abl tyrosine kinase; Abelsone tyrosine kinase (abl) | |
| 2 | View Details | [416..484] | 2.1 | Regulatory apparatus of Calcium Dependent protein kinase from Arabidopsis thaliana | |
| 3 | View Details | [485..599] | 13.30103 | Calcineurin regulatory subunit (B-chain) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)