













| General Information: |
|
| Name(s) found: |
CSP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 299 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to cold
[IGI]
DNA duplex unwinding [IDA] RNA secondary structure unwinding [IDA] |
| Molecular Function: |
nucleic acid binding
[ISS]
RNA binding [IDA] double-stranded DNA binding [IDA] single-stranded DNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASEDQSAAR STGKVNWFNA SKGYGFITPD DGSVELFVHQ SSIVSEGYRS LTVGDAVEFA 60
61 ITQGSDGKTK AVNVTAPGGG SLKKENNSRG NGARRGGGGS GCYNCGELGH ISKDCGIGGG 120
121 GGGGERRSRG GEGCYNCGDT GHFARDCTSA GNGDQRGATK GGNDGCYTCG DVGHVARDCT 180
181 QKSVGNGDQR GAVKGGNDGC YTCGDVGHFA RDCTQKVAAG NVRSGGGGSG TCYSCGGVGH 240
241 IARDCATKRQ PSRGCYQCGG SGHLARDCDQ RGSGGGGNDN ACYKCGKEGH FARECSSVA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
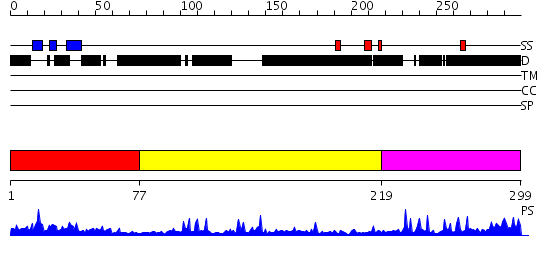
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..76] | 18.39794 | CRYSTAL STRUCTURE OF CSPA, THE MAJOR COLD SHOCK PROTEIN OF ESCHERICHIA COLI | |
| 2 | View Details | [77..218] | 15.045757 | HIV-1 CAPSID PROTEIN C-TERMINAL FRAGMENT PLUS GAG P2 DOMAIN | |
| 3 | View Details | [219..299] | 8.221849 | NUCLEOCAPSID ZINC FINGERS DETECTED IN RETROVIRUSES: EXAFS STUDIES ON INTACT VIRUSES AND THE SOLUTION-STATE STRUCTURE OF THE NUCLEOCAPSID PROTEIN FROM HIV-1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)