













| General Information: |
|
| Name(s) found: |
ASE2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 561 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
plastid stroma [IDA] |
| Biological Process: |
purine nucleotide biosynthetic process
[IMP][TAS]
purine base biosynthetic process [ISS] leaf morphogenesis [IMP] |
| Molecular Function: |
amidophosphoribosyltransferase activity
[IMP][ISS][TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAATSSISSS LSLNAKPNKL SNNNNNNKPH RFLRNPFLNP SSSSFSPLPA SISSSSSSPS 60
61 FPLRVSNPLT LLAADNDDYD EKPREECGVV GIYGDSEASR LCYLALHALQ HRGQEGAGIV 120
121 TVSKDKVLQT ITGVGLVSEV FSESKLDQLP GDIAIGHVRY STAGSSMLKN VQPFVAGYRF 180
181 GSVGVAHNGN LVNYTKLRAD LEENGSIFNT SSDTEVVLHL IAISKARPFF MRIVDACEKL 240
241 QGAYSMVFVT EDKLVAVRDP HGFRPLVMGR RSNGAVVFAS ETCALDLIEA TYEREVYPGE 300
301 VLVVDKDGVK CQCLMPHPEP KQCIFEHIYF SLPNSIVFGR SVYESRHVFG EILATESPVD 360
361 CDVVIAVPDS GVVAALGYAA KAGVAFQQGL IRSHYVGRTF IEPSQKIRDF GVKLKLSPVR 420
421 GVLEGKRVVV VDDSIVRGTT SSKIVRLLRE AGAKEVHMRI ASPPIIASCY YGVDTPSSNE 480
481 LISNRMSVDE IRDYIGCDSL AFLSFETLKK HLGEDSRSFC YACFTGDYPV KPTEDKVKRG 540
541 GDFIDDGLVG GIHNIEGGWV R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
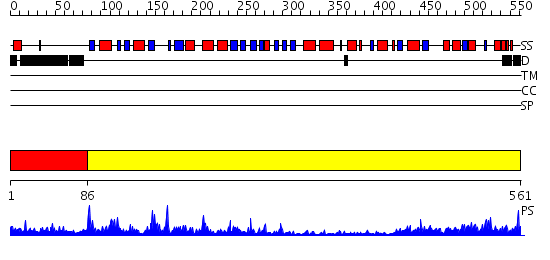
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | 43.69897 | Alpha subunit of glutamate synthase, C-terminal domain; Alpha subunit of glutamate synthase, central and FMN domains; Alpha subunit of glutamate synthase, N-terminal domain | |
| 2 | View Details | [86..561] | 118.0 | Glutamine PRPP amidotransferase, C-terminal domain; Glutamine PRPP amidotransferase, N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)