













| General Information: |
|
| Name(s) found: |
DRP3A_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 808 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[IDA]
chloroplast [IDA] membrane [IDA] mitochondrion [IDA] chloroplast envelope [IDA] cytosol [IDA] |
| Biological Process: |
peroxisome fission
[IMP]
post-embryonic development [IGI] chloroplast organization [NAS] mitochondrial fission [IGI] |
| Molecular Function: |
GTP binding
[ISS]
GTPase activity [IDA][ISS] phosphoinositide binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTIEEVSGET PPSTPPSSST PSPSSSTTNA APLGSSVIPI VNKLQDIFAQ LGSQSTIALP 60
61 QVVVVGSQSS GKSSVLEALV GRDFLPRGND ICTRRPLVLQ LLQTKSRANG GSDDEWGEFR 120
121 HLPETRFYDF SEIRREIEAE TNRLVGENKG VADTQIRLKI SSPNVLNITL VDLPGITKVP 180
181 VGDQPSDIEA RIRTMILSYI KQDTCLILAV TPANTDLANS DALQIASIVD PDGHRTIGVI 240
241 TKLDIMDKGT DARKLLLGNV VPLRLGYVGV VNRCQEDILL NRTVKEALLA EEKFFRSHPV 300
301 YHGLADRLGV PQLAKKLNQI LVQHIKVLLP DLKSRISNAL VATAKEHQSY GELTESRAGQ 360
361 GALLLNFLSK YCEAYSSLLE GKSEEMSTSE LSGGARIHYI FQSIFVKSLE EVDPCEDLTD 420
421 DDIRTAIQNA TGPRSALFVP DVPFEVLVRR QISRLLDPSL QCARFIFEEL IKISHRCMMN 480
481 ELQRFPVLRK RMDEVIGDFL REGLEPSEAM IGDIIDMEMD YINTSHPNFI GGTKAVEAAM 540
541 HQVKSSRIPH PVARPKDTVE PDRTSSSTSQ VKSRSFLGRQ ANGIVTDQGV VSADAEKAQP 600
601 AANASDTRWG IPSIFRGGDT RAVTKDSLLN KPFSEAVEDM SHNLSMIYLK EPPAVLRPTE 660
661 THSEQEAVEI QITKLLLRSY YDIVRKNIED SVPKAIMHFL VNHTKRELHN VFIKKLYREN 720
721 LFEEMLQEPD EIAVKRKRTQ ETLHVLQQAY RTLDELPLEA DSVSAGMSKH QELLTSSKYS 780
781 TSSSYSASPS TTRRSRRAGD QHQNGYGF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
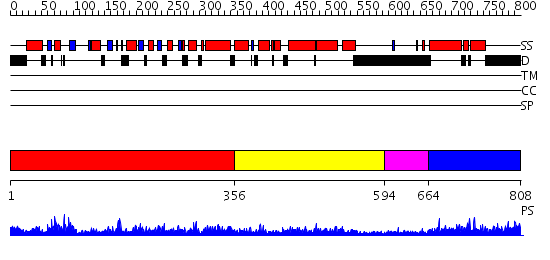
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..355] | 103.0 | Dynamin G domain | |
| 2 | View Details | [356..593] | 50.408935 | No description for PF01031.8 was found. No confident structure predictions are available. | |
| 3 | View Details | [594..663] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [664..808] | 43.30103 | No description for PF02212.6 was found. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)