













| General Information: |
|
| Name(s) found: |
CPNB4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 611 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
cellular protein metabolic process
[IEA]
|
| Molecular Function: |
ATP binding
[IEA]
protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAFSQAALSA LPLSDRTFRK KPSSSSSSSP NFVLRVRAAA KEVHFNRDGS VTKKLQAGAD 60
61 MVAKLLGVTL GPKGRNVVLQ NKYGPPRIVN DGETVLKEIE LEDPLENVGV KLVRQAGAKT 120
121 NDLAGDGSTT SIILAHGLIT EGIKVISAGT NPIQVARGIE KTTKALVLEL KSMSREIEDH 180
181 ELAHVAAVSA GNDYEVGNMI SNAFQQVGRT GVVTIEKGKY LVNNLEIVEG MQFNRGYLSP 240
241 YFVTDRRKRE AEFHDCKLLL VDKKITNPKD MFKILDSAVK EEFPVLIVAE DIEQDALAPV 300
301 IRNKLKGNLK VAAIKAPAFG ERKSHCLDDL AIFTGATVIR DEMGLSLEKA GKEVLGTAKR 360
361 VLVTKDSTLI VTNGFTQKAV DERVSQIKNL IENTEENFQK KILNERVARL SGGIAIIQVG 420
421 ALTQVELKDK QLKVEDALNA TKSAIEEGIV VGGGCALLRL ATKVDRIKET LDNTEQKIGA 480
481 EIFKKALSYP IRLIAKNADT NGNIVIEKVL SNKNTMYGYN AAKNQYEDLM LAGIIDPTKV 540
541 VRCCLEHASS VAQTFLTSDC VVVEIKEIKP RPIINPPLPT SSPATSSMFP DRKLPRFPQI 600
601 MPRTRSHFPR K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
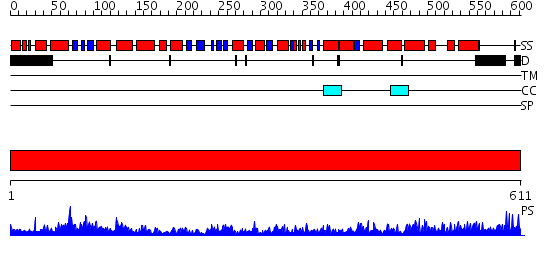
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..611] | 169.0 | Crystal Structure of the Chaperonin Complex Cpn60/Cpn10/(ADP)7 from Thermus Thermophilus |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.63 |
Source: Reynolds et al. (2008)