













| General Information: |
|
| Name(s) found: |
VATA_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 623 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
cell wall [IDA] membrane [IDA] chloroplast envelope [IDA] plant-type vacuole [IDA] vacuolar membrane [IDA] vacuole [IDA] plasma membrane [IDA] |
| Biological Process: |
proton transport
[ISS]
response to salt stress [IEP] Golgi organization [IMP] pollen development [IMP] |
| Molecular Function: |
ATP binding
[ISS]
proton-transporting ATPase activity, rotational mechanism [IEA] hydrogen ion transporting ATP synthase activity, rotational mechanism [IEA][ISS] hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPAFYGGKLT TFEDDEKESE YGYVRKVSGP VVVADGMAGA AMYELVRVGH DNLIGEIIRL 60
61 EGDSATIQVY EETAGLTVND PVLRTHKPLS VELGPGILGN IFDGIQRPLK TIARISGDVY 120
121 IPRGVSVPAL DKDCLWEFQP NKFVEGDTIT GGDLYATVFE NTLMNHLVAL PPDAMGKITY 180
181 IAPAGQYSLK DTVIELEFQG IKKSYTMLQS WPVRTPRPVA SKLAADTPLL TGQRVLDALF 240
241 PSVLGGTCAI PGAFGCGKTV ISQALSKYSN SDAVVYVGCG ERGNEMAEVL MDFPQLTMTL 300
301 PDGREESVMK RTTLVANTSN MPVAAREASI YTGITIAEYF RDMGYNVSMM ADSTSRWAEA 360
361 LREISGRLAE MPADSGYPAY LAARLASFYE RAGKVKCLGG PERNGSVTIV GAVSPPGGDF 420
421 SDPVTSATLS IVQVFWGLDK KLAQRKHFPS VNWLISYSKY STALESFYEK FDPDFINIRT 480
481 KAREVLQRED DLNEIVQLVG KDALAEGDKI TLETAKLLRE DYLAQNAFTP YDKFCPFYKS 540
541 VWMMRNIIHF YNLANQAVER AAGMDGQKIT YTLIKHRLGD LFYRLVSQKF EDPAEGEDTL 600
601 VEKFKKLYDD LNAGFRALED ETR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
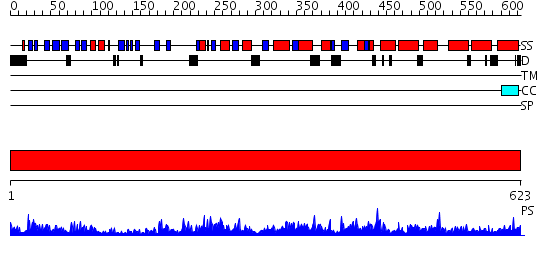
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..623] | 154.0 | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)